You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001183_01738
You are here: Home > Sequence: MGYG000001183_01738
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
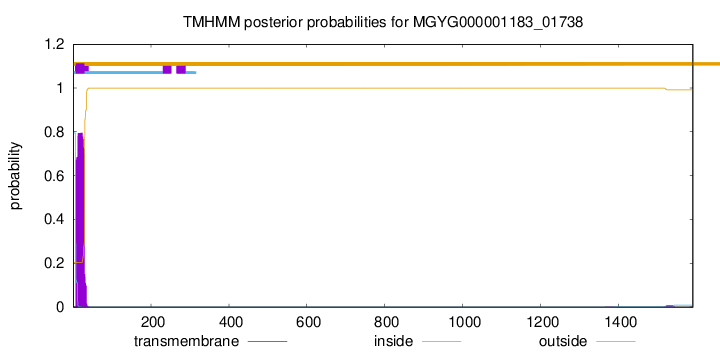
TMHMM annotations
Basic Information help
| Species | UMGS1441 sp900551755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; CAG-274; UMGS1441; UMGS1441 sp900551755 | |||||||||||
| CAZyme ID | MGYG000001183_01738 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16674; End: 21449 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 246 | 434 | 1.5e-37 | 0.8465346534653465 |
| CBM77 | 1477 | 1583 | 9e-36 | 0.9902912621359223 |
| CBM77 | 796 | 894 | 2e-18 | 0.9320388349514563 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18283 | CBM77 | 1.39e-35 | 1475 | 1585 | 1 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| COG3866 | PelB | 7.44e-26 | 251 | 437 | 98 | 278 | Pectate lyase [Carbohydrate transport and metabolism]. |
| smart00656 | Amb_all | 1.87e-19 | 255 | 436 | 17 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 2.38e-11 | 290 | 432 | 64 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| cd14256 | Dockerin_I | 7.09e-06 | 1111 | 1165 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBF42492.1 | 4.20e-135 | 34 | 534 | 39 | 504 |
| ADU23076.1 | 9.78e-94 | 35 | 532 | 761 | 1237 |
| CDR31241.1 | 1.98e-88 | 35 | 537 | 333 | 812 |
| ACR71161.1 | 2.59e-85 | 37 | 892 | 47 | 882 |
| ADZ82421.1 | 1.30e-66 | 1 | 660 | 2 | 618 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FU5_A | 1.87e-29 | 1474 | 1587 | 5 | 113 | Thecomplexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition [Ruminococcus flavefaciens] |
| 3VMV_A | 1.51e-17 | 254 | 433 | 79 | 247 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1VBL_A | 1.32e-15 | 253 | 432 | 131 | 330 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 2QX3_A | 8.09e-14 | 290 | 445 | 94 | 259 | Structureof pectate lyase II from Xanthomonas campestris pv. campestris str. ATCC 33913 [Xanthomonas campestris pv. campestris],2QX3_B Structure of pectate lyase II from Xanthomonas campestris pv. campestris str. ATCC 33913 [Xanthomonas campestris pv. campestris] |
| 2QY1_A | 1.45e-13 | 290 | 420 | 94 | 234 | ChainA, Pectate lyase II [Xanthomonas campestris pv. campestris],2QY1_B Chain B, Pectate lyase II [Xanthomonas campestris pv. campestris] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1C2 | 1.17e-15 | 247 | 451 | 103 | 297 | Pectate lyase 3 OS=Pectobacterium carotovorum OX=554 GN=pel3 PE=1 SV=1 |
| P0C1C3 | 1.17e-15 | 247 | 451 | 103 | 297 | Pectate lyase 3 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=pel3 PE=3 SV=1 |
| P0C1C0 | 2.79e-15 | 290 | 451 | 136 | 297 | Pectate lyase 1 OS=Pectobacterium carotovorum OX=554 GN=pel1 PE=1 SV=1 |
| Q6CZT2 | 8.92e-15 | 211 | 451 | 71 | 297 | Pectate lyase 3 OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=pel3 PE=3 SV=1 |
| Q6CZT4 | 2.13e-14 | 290 | 451 | 136 | 297 | Pectate lyase 1 OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=pel1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000378 | 0.998751 | 0.000200 | 0.000257 | 0.000189 | 0.000171 |