You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001186_02705
You are here: Home > Sequence: MGYG000001186_02705
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Agathobacter sp900547695 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Agathobacter; Agathobacter sp900547695 | |||||||||||
| CAZyme ID | MGYG000001186_02705 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 32203; End: 33471 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 134 | 373 | 4.5e-93 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.39e-69 | 131 | 381 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.71e-07 | 155 | 316 | 74 | 276 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG3867 | GanB | 4.80e-04 | 147 | 240 | 55 | 137 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SIP56455.1 | 1.01e-168 | 99 | 422 | 63 | 386 |
| ACR74405.1 | 3.10e-168 | 99 | 422 | 63 | 386 |
| CBK89462.1 | 9.50e-168 | 99 | 422 | 67 | 390 |
| CBK94241.1 | 1.66e-167 | 99 | 422 | 63 | 386 |
| SIP56505.1 | 9.53e-167 | 99 | 422 | 63 | 386 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 5.19e-88 | 121 | 409 | 11 | 296 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 4XZW_A | 7.23e-85 | 120 | 414 | 9 | 303 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 4.20e-84 | 120 | 414 | 9 | 304 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 3PZT_A | 5.02e-83 | 120 | 409 | 34 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 1H5V_A | 7.71e-79 | 114 | 409 | 3 | 296 | Thiopentasaccharidecomplex of the endoglucanase Cel5A from Bacillus agaradharens at 1.1 A resolution in the tetragonal crystal form [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22541 | 9.53e-104 | 121 | 409 | 114 | 403 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| Q07940 | 2.33e-91 | 122 | 407 | 4 | 288 | Endoglucanase 4 OS=Ruminococcus albus OX=1264 GN=Eg IV PE=1 SV=1 |
| P15704 | 3.70e-87 | 115 | 406 | 39 | 328 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P10475 | 3.79e-80 | 120 | 409 | 39 | 325 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P07983 | 1.49e-79 | 120 | 409 | 39 | 325 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
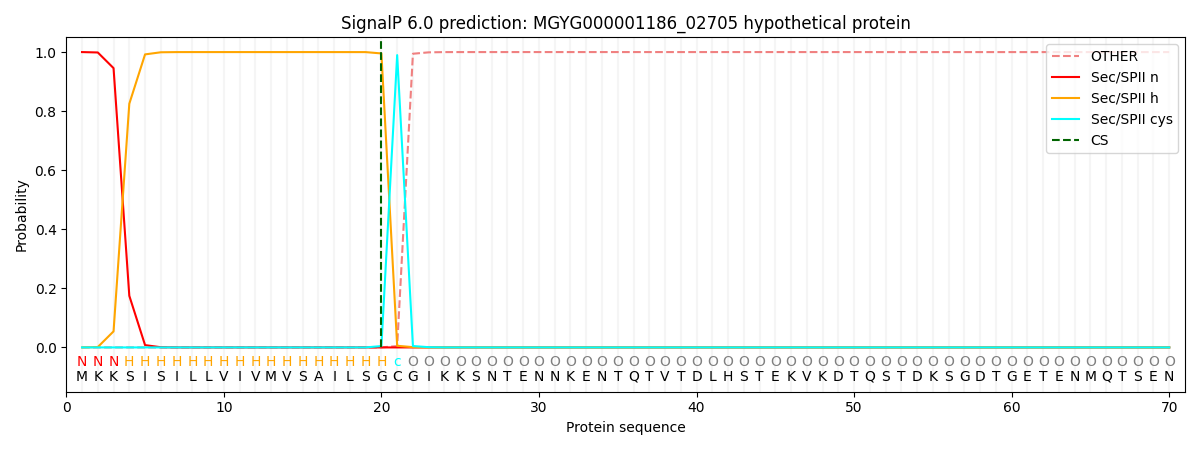
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000083 | 0.000000 | 0.000000 | 0.000000 |
