You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001210_01296
You are here: Home > Sequence: MGYG000001210_01296
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
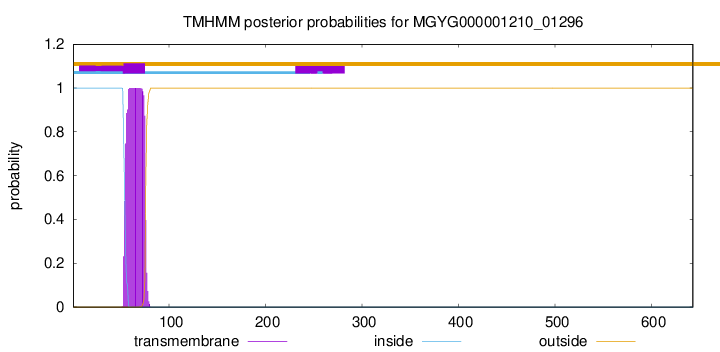
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter sp900553885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter sp900553885 | |||||||||||
| CAZyme ID | MGYG000001210_01296 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 931; End: 2862 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 326 | 631 | 7.7e-37 | 0.7871621621621622 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02874 | GH18_CFLE_spore_hydrolase | 3.33e-65 | 325 | 638 | 4 | 313 | Cortical fragment-lytic enzyme (CFLE) is a peptidoglycan hydrolase involved in bacterial endospore germination. CFLE is expressed as an inactive preprotein (called SleB) in the forespore compartment of sporulating cells. SleB translocates across the forespore inner membrane and is deposited as a mature enzyme in the cortex layer of the spore. As part of a sensory mechanism capable of initiating germination, CFLE degrades a spore-specific peptidoglycan constituent called muramic-acid delta-lactam that comprises the outer cortex. CFLE has a C-terminal glycosyl hydrolase family 18 (GH18) catalytic domain as well as two N-terminal LysM peptidoglycan-binding domains. In addition to SleB, this family includes YaaH, YdhD, and YvbX from Bacillus subtilis. |
| COG3858 | YaaH | 3.32e-39 | 328 | 638 | 116 | 418 | Spore germination protein YaaH [Cell cycle control, cell division, chromosome partitioning]. |
| smart00636 | Glyco_18 | 1.40e-28 | 384 | 629 | 74 | 333 | Glyco_18 domain. |
| cd06549 | GH18_trifunctional | 1.48e-27 | 339 | 635 | 24 | 298 | GH18 domain of an uncharacterized family of bacterial proteins, which share a common three-domain architecture: an N-terminal glycosyl hydrolase family 18 (GH18) domain, a glycosyl transferase family 2 domain, and a C-terminal polysaccharide deacetylase domain. |
| pfam00704 | Glyco_hydro_18 | 2.09e-27 | 384 | 629 | 71 | 306 | Glycosyl hydrolases family 18. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT30243.1 | 3.86e-287 | 75 | 643 | 83 | 645 |
| QEI31514.1 | 7.77e-287 | 75 | 643 | 83 | 645 |
| QHB24012.1 | 7.77e-287 | 75 | 643 | 83 | 645 |
| CBL24889.1 | 1.37e-279 | 125 | 643 | 134 | 652 |
| QEK19554.1 | 7.51e-243 | 74 | 643 | 109 | 664 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CZ8_A | 1.61e-22 | 315 | 631 | 11 | 312 | ChainA, Putative sporulation-specific glycosylase ydhD [Bacillus subtilis subsp. subtilis str. 168],3CZ8_B Chain B, Putative sporulation-specific glycosylase ydhD [Bacillus subtilis subsp. subtilis str. 168] |
| 4S3J_A | 1.75e-17 | 366 | 640 | 160 | 423 | Crystalstructure of the Bacillus cereus spore cortex-lytic enzyme SleL [Bacillus cereus ATCC 10876],4S3J_B Crystal structure of the Bacillus cereus spore cortex-lytic enzyme SleL [Bacillus cereus ATCC 10876],4S3J_C Crystal structure of the Bacillus cereus spore cortex-lytic enzyme SleL [Bacillus cereus ATCC 10876] |
| 4S3K_A | 1.31e-15 | 337 | 638 | 133 | 423 | ChainA, Spore germination protein YaaH [Priestia megaterium QM B1551] |
| 4MNJ_A | 7.20e-09 | 384 | 630 | 78 | 333 | CrystalStructure of GH18 Chitinase from Cycad, Cycas revoluta [Cycas revoluta],4MNK_A Crystal Structure of GH18 Chitinase from Cycas revoluta in complex with (GlcNAc)3 [Cycas revoluta],4R5E_A Chain A, Chitinase A [Cycas revoluta] |
| 6BT9_A | 1.38e-08 | 416 | 627 | 197 | 445 | ChitinaseChiA74 from Bacillus thuringiensis [Bacillus thuringiensis],6BT9_B Chitinase ChiA74 from Bacillus thuringiensis [Bacillus thuringiensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O32258 | 5.30e-22 | 339 | 638 | 52 | 339 | Uncharacterized glycosylase YvbX OS=Bacillus subtilis (strain 168) OX=224308 GN=yvbX PE=3 SV=1 |
| O05495 | 1.38e-21 | 315 | 627 | 103 | 400 | Putative sporulation-specific glycosylase YdhD OS=Bacillus subtilis (strain 168) OX=224308 GN=ydhD PE=1 SV=2 |
| P37531 | 3.97e-16 | 395 | 643 | 186 | 424 | Cortical fragment-lytic enzyme OS=Bacillus subtilis (strain 168) OX=224308 GN=sleL PE=1 SV=2 |
| P0DPJ9 | 9.34e-16 | 366 | 640 | 159 | 422 | Cortical fragment-lytic enzyme OS=Bacillus anthracis OX=1392 GN=sleL PE=2 SV=1 |
| Q9K3E4 | 2.94e-15 | 366 | 640 | 159 | 422 | Cortical fragment-lytic enzyme OS=Bacillus cereus OX=1396 GN=sleL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
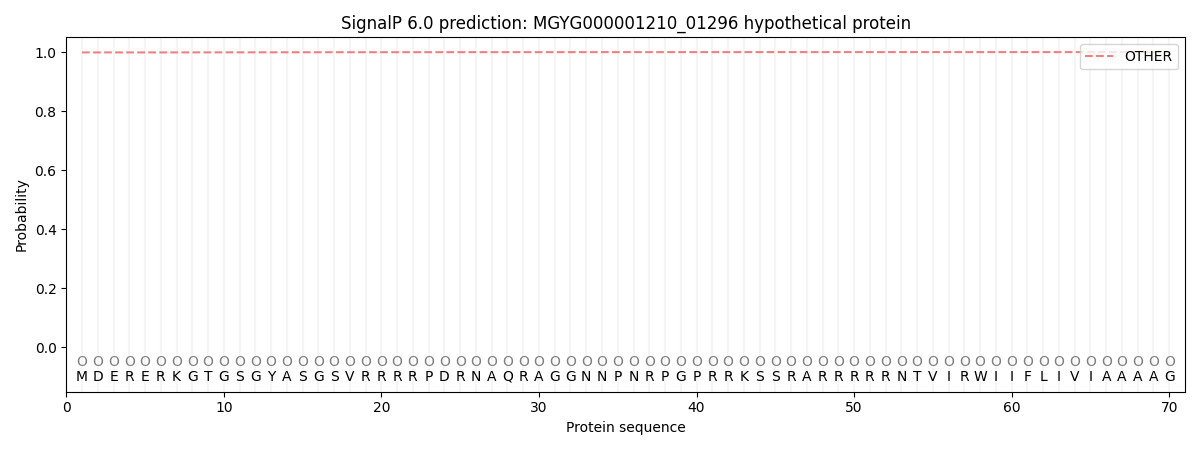
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999223 | 0.000765 | 0.000000 | 0.000000 | 0.000000 | 0.000001 |