You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001220_01079
You are here: Home > Sequence: MGYG000001220_01079
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UMGS1820 sp900555375 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales; Monoglobaceae; UMGS1820; UMGS1820 sp900555375 | |||||||||||
| CAZyme ID | MGYG000001220_01079 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3875; End: 5677 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 274 | 544 | 1.2e-65 | 0.9923954372623575 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.11e-26 | 290 | 554 | 20 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.09e-22 | 233 | 564 | 11 | 335 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00395 | SLH | 7.69e-10 | 89 | 125 | 1 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 4.09e-07 | 150 | 192 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 5.88e-06 | 84 | 191 | 638 | 743 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ98812.1 | 1.69e-104 | 224 | 594 | 1 | 363 |
| AQR93159.1 | 7.49e-97 | 224 | 594 | 60 | 431 |
| AVM42141.1 | 3.99e-94 | 242 | 600 | 4 | 358 |
| BCJ97694.1 | 3.49e-61 | 221 | 575 | 45 | 412 |
| QAA34430.1 | 1.32e-57 | 232 | 574 | 49 | 380 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1CEC_A | 4.00e-17 | 287 | 489 | 21 | 201 | ChainA, ENDOGLUCANASE CELC [Acetivibrio thermocellus] |
| 1CEN_A | 9.67e-17 | 287 | 489 | 21 | 201 | ChainA, CELLULASE CELC [Acetivibrio thermocellus],1CEO_A Chain A, CELLULASE CELC [Acetivibrio thermocellus] |
| 1H4P_A | 5.25e-12 | 230 | 552 | 3 | 317 | Crystalstructure of exo-1,3-beta glucanse from Saccharomyces cerevisiae [Saccharomyces cerevisiae],1H4P_B Crystal structure of exo-1,3-beta glucanse from Saccharomyces cerevisiae [Saccharomyces cerevisiae] |
| 3AMC_A | 1.65e-11 | 295 | 583 | 34 | 316 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
| 6ZB9_A | 6.28e-11 | 240 | 416 | 3 | 168 | ChainA, Exo-beta-1,3-glucanase [uncultured bacterium],6ZB9_B Chain B, Exo-beta-1,3-glucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 2.58e-27 | 225 | 552 | 10 | 344 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| Q8NKF9 | 9.70e-21 | 236 | 416 | 33 | 215 | Glucan 1,3-beta-glucosidase OS=Candida oleophila OX=45573 GN=EXG1 PE=3 SV=1 |
| Q12626 | 8.45e-18 | 237 | 416 | 46 | 223 | Glucan 1,3-beta-glucosidase OS=Pichia angusta OX=870730 PE=3 SV=1 |
| A1D4Q5 | 1.75e-17 | 237 | 421 | 39 | 217 | Probable glucan 1,3-beta-glucosidase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=exgA PE=3 SV=1 |
| A2RAR6 | 2.34e-17 | 240 | 423 | 42 | 218 | Probable glucan 1,3-beta-glucosidase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=exgA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
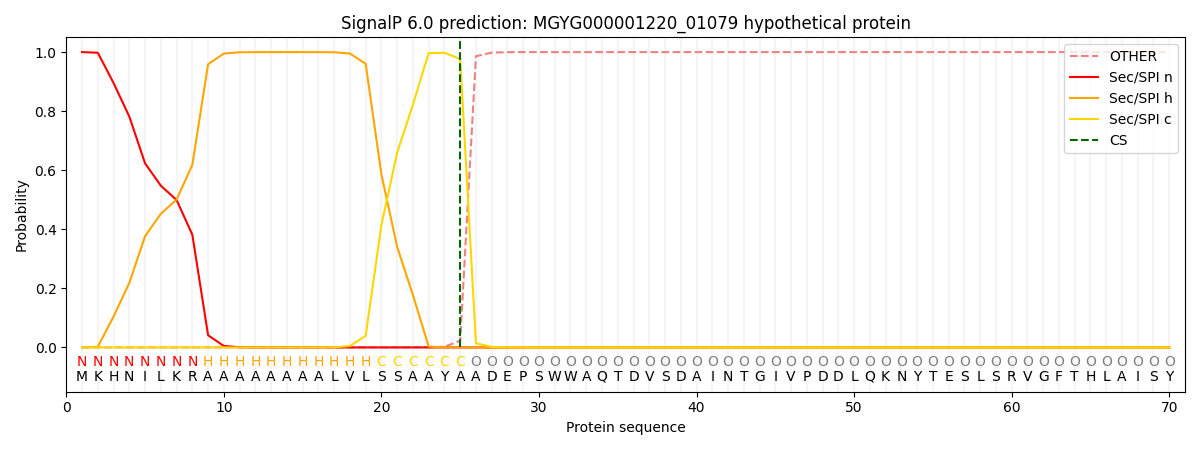
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000615 | 0.998245 | 0.000269 | 0.000318 | 0.000297 | 0.000246 |
