You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001230_01256
You are here: Home > Sequence: MGYG000001230_01256
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
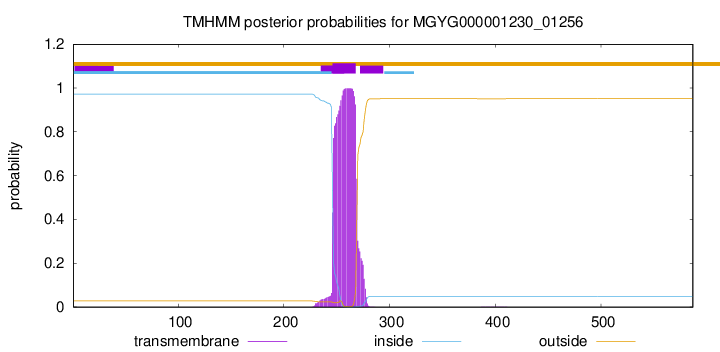
TMHMM annotations
Basic Information help
| Species | Eubacterium_R sp900542875 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Acutalibacteraceae; Eubacterium_R; Eubacterium_R sp900542875 | |||||||||||
| CAZyme ID | MGYG000001230_01256 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 668; End: 2431 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16891 | CwlT-like | 2.30e-54 | 294 | 438 | 1 | 150 | CwlT-like N-terminal lysozyme domain and similar domains. CwlT is a bifunctional cell wall hydrolase containing an N-terminal lysozyme domain and a C-terminal NlpC/P60 endopeptidase domain (gamma-d-D-glutamyl-L-diamino acid endopeptidase), and has been implicated in the spread of transposons. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam13702 | Lysozyme_like | 6.79e-47 | 288 | 454 | 1 | 159 | Lysozyme-like. |
| pfam05257 | CHAP | 7.51e-18 | 472 | 562 | 1 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
| cd13401 | Slt70-like | 2.10e-05 | 293 | 402 | 5 | 120 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| cd16896 | LT_Slt70-like | 1.54e-04 | 294 | 438 | 4 | 144 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO05014.1 | 0.0 | 1 | 587 | 1 | 591 |
| AIK73318.1 | 0.0 | 1 | 587 | 1 | 591 |
| AIK22064.1 | 0.0 | 1 | 587 | 1 | 591 |
| QOZ88918.1 | 0.0 | 1 | 587 | 1 | 591 |
| VCV20591.1 | 0.0 | 1 | 587 | 1 | 591 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FDY_A | 4.13e-15 | 287 | 534 | 22 | 261 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
| 4HPE_A | 1.97e-11 | 287 | 547 | 20 | 272 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96645 | 9.00e-36 | 276 | 547 | 42 | 293 | Probable endopeptidase YddH OS=Bacillus subtilis (strain 168) OX=224308 GN=yddH PE=3 SV=1 |
| O34636 | 5.24e-11 | 285 | 419 | 46 | 171 | Uncharacterized membrane protein YocA OS=Bacillus subtilis (strain 168) OX=224308 GN=yocA PE=4 SV=1 |
| Q5XC63 | 3.19e-09 | 291 | 449 | 30 | 179 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M6 (strain ATCC BAA-946 / MGAS10394) OX=286636 GN=pvaA PE=3 SV=1 |
| Q8P120 | 3.19e-09 | 291 | 449 | 30 | 179 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M18 (strain MGAS8232) OX=186103 GN=pvaA PE=3 SV=1 |
| Q99ZN9 | 3.19e-09 | 291 | 449 | 30 | 179 | Pneumococcal vaccine antigen A homolog OS=Streptococcus pyogenes serotype M1 OX=301447 GN=pvaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
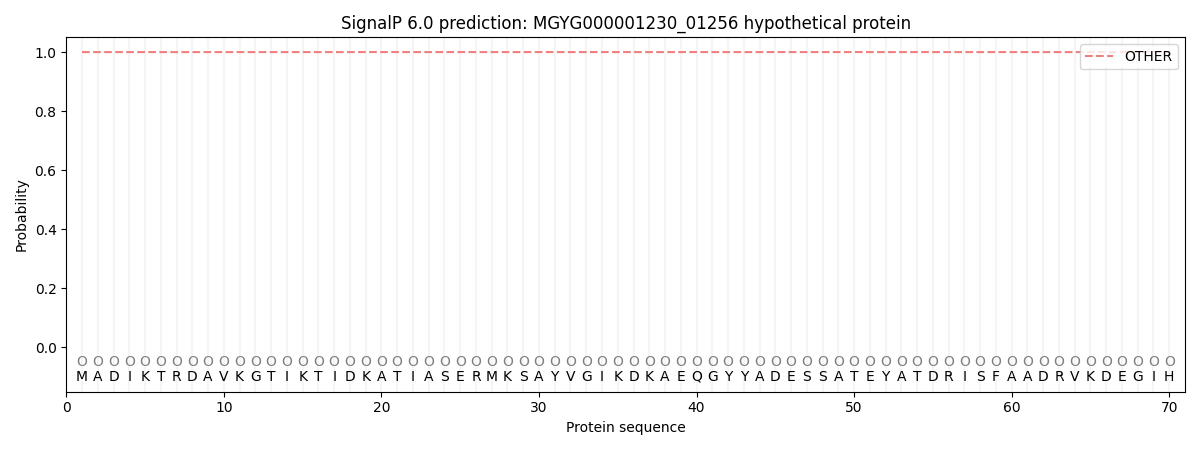
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000042 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |