You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001232_00890
You are here: Home > Sequence: MGYG000001232_00890
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
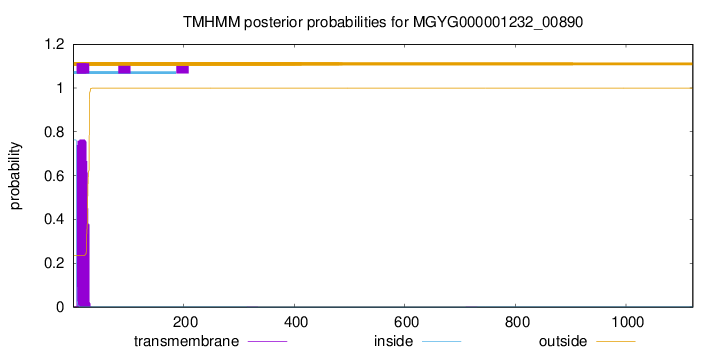
TMHMM annotations
Basic Information help
| Species | Peptacetobacter sp900550335 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Peptacetobacter; Peptacetobacter sp900550335 | |||||||||||
| CAZyme ID | MGYG000001232_00890 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 142; End: 3507 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 42 | 373 | 1e-51 | 0.884272997032641 |
| GH20 | 470 | 806 | 7.5e-45 | 0.8961424332344213 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06564 | GH20_DspB_LnbB-like | 1.71e-86 | 469 | 818 | 4 | 323 | Glycosyl hydrolase family 20 (GH20) catalytic domain of dispersin B (DspB), lacto-N-biosidase (LnbB) and related proteins. Dispersin B is a soluble beta-N-acetylglucosamidase found in bacteria that hydrolyzes the beta-1,6-linkages of PGA (poly-beta-(1,6)-N-acetylglucosamine), a major component of the extracellular polysaccharide matrix. Lacto-N-biosidase hydrolyzes lacto-N-biose (LNB) type I oligosaccharides at the nonreducing terminus to produce lacto-N-biose as part of the GNB/LNB (galacto-N-biose/lacto-N-biose I) degradation pathway. The lacto-N-biosidase from Bifidobacterium bifidum has this GH20 domain, a carbohydrate binding module 32, and a bacterial immunoglobulin-like domain 2, as well as a YSIRK signal peptide and a G5 membrane anchor at the N and C termini, respectively. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| cd06564 | GH20_DspB_LnbB-like | 1.25e-84 | 38 | 387 | 1 | 321 | Glycosyl hydrolase family 20 (GH20) catalytic domain of dispersin B (DspB), lacto-N-biosidase (LnbB) and related proteins. Dispersin B is a soluble beta-N-acetylglucosamidase found in bacteria that hydrolyzes the beta-1,6-linkages of PGA (poly-beta-(1,6)-N-acetylglucosamine), a major component of the extracellular polysaccharide matrix. Lacto-N-biosidase hydrolyzes lacto-N-biose (LNB) type I oligosaccharides at the nonreducing terminus to produce lacto-N-biose as part of the GNB/LNB (galacto-N-biose/lacto-N-biose I) degradation pathway. The lacto-N-biosidase from Bifidobacterium bifidum has this GH20 domain, a carbohydrate binding module 32, and a bacterial immunoglobulin-like domain 2, as well as a YSIRK signal peptide and a G5 membrane anchor at the N and C termini, respectively. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| COG3525 | Chb | 8.94e-75 | 43 | 810 | 16 | 732 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
| pfam04122 | CW_binding_2 | 1.25e-23 | 834 | 917 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
| pfam04122 | CW_binding_2 | 4.22e-23 | 1033 | 1118 | 1 | 80 | Putative cell wall binding repeat 2. This repeat is found in multiple tandem copies in proteins including amidase enhancers and adhesins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK21709.1 | 0.0 | 1 | 1120 | 1 | 1120 |
| QQQ86831.1 | 0.0 | 1 | 1120 | 1 | 1120 |
| AQA08098.1 | 7.88e-243 | 19 | 823 | 137 | 942 |
| VFI19059.1 | 1.39e-241 | 38 | 823 | 168 | 962 |
| QXW61492.1 | 4.79e-241 | 38 | 823 | 158 | 942 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7PUL_A | 1.66e-126 | 468 | 822 | 11 | 350 | ChainA, Beta-N-acetylhexosaminidase [Enterococcus faecalis] |
| 4AZI_A | 1.43e-120 | 38 | 462 | 9 | 424 | Differentialinhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH [Streptococcus pneumoniae TIGR4],4AZI_B Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH [Streptococcus pneumoniae TIGR4] |
| 2YLL_A | 8.05e-120 | 23 | 431 | 11 | 421 | Inhibitionof the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis [Streptococcus pneumoniae TIGR4] |
| 4AZ7_A | 5.95e-119 | 466 | 821 | 6 | 364 | Differentialinhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH [Streptococcus pneumoniae TIGR4] |
| 2YL6_A | 6.15e-119 | 466 | 821 | 6 | 364 | Inhibitionof the pneumococcal virulence factor StrH and molecular insights into N-glycan recognition and hydrolysis [Streptococcus pneumoniae TIGR4],4AZ5_A Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49610 | 1.44e-234 | 38 | 823 | 186 | 980 | Beta-N-acetylhexosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=strH PE=1 SV=2 |
| Q02114 | 5.27e-41 | 830 | 1121 | 26 | 311 | N-acetylmuramoyl-L-alanine amidase LytC OS=Bacillus subtilis (strain 168) OX=224308 GN=lytC PE=1 SV=1 |
| Q02113 | 1.62e-14 | 928 | 1121 | 57 | 242 | Amidase enhancer OS=Bacillus subtilis (strain 168) OX=224308 GN=lytB PE=1 SV=1 |
| P13723 | 3.19e-07 | 29 | 316 | 147 | 398 | Beta-hexosaminidase subunit A1 OS=Dictyostelium discoideum OX=44689 GN=hexa1 PE=1 SV=1 |
| Q9UYS2 | 3.03e-06 | 962 | 1090 | 30 | 166 | Uncharacterized protein PYRAB14350 OS=Pyrococcus abyssi (strain GE5 / Orsay) OX=272844 GN=PYRAB14350 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
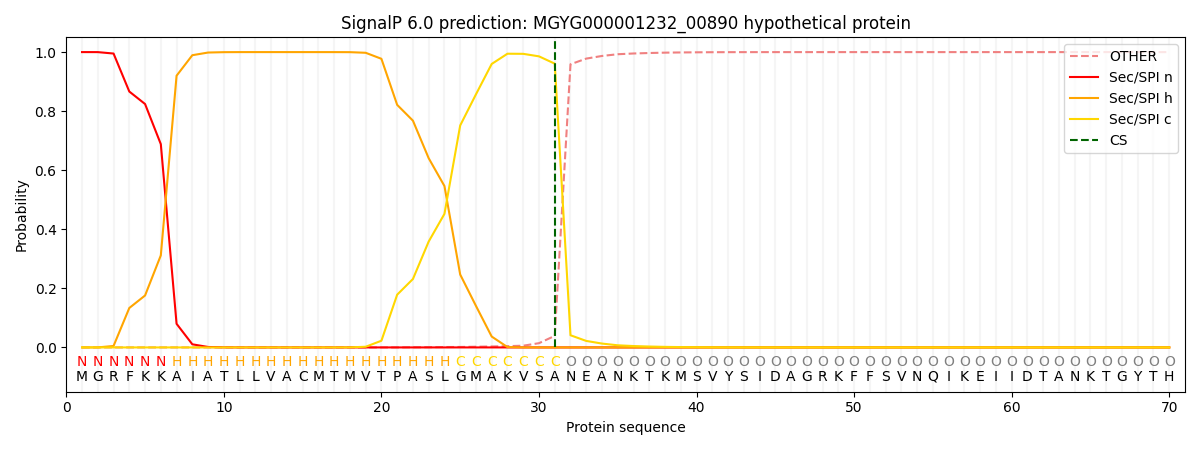
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000253 | 0.999072 | 0.000192 | 0.000177 | 0.000150 | 0.000137 |