You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001240_00379
You are here: Home > Sequence: MGYG000001240_00379
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
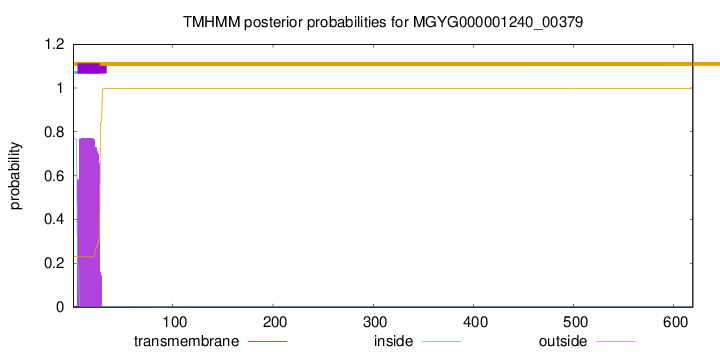
TMHMM annotations
Basic Information help
| Species | Prevotella sp900557405 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900557405 | |||||||||||
| CAZyme ID | MGYG000001240_00379 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 53770; End: 55629 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 23 | 582 | 3.1e-190 | 0.9943074003795066 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 4.98e-10 | 405 | 548 | 8 | 112 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.04e-06 | 374 | 548 | 1 | 136 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 0.001 | 375 | 499 | 25 | 147 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05048 | NosD | 0.005 | 369 | 544 | 37 | 170 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD42515.1 | 7.53e-280 | 8 | 602 | 7 | 602 |
| ALK85163.1 | 2.77e-275 | 1 | 611 | 46 | 654 |
| QQY37289.1 | 3.17e-275 | 1 | 611 | 1 | 609 |
| QEW37044.1 | 8.42e-275 | 1 | 611 | 38 | 646 |
| QQY43633.1 | 1.28e-274 | 1 | 611 | 1 | 609 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 5.67e-170 | 20 | 612 | 23 | 615 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
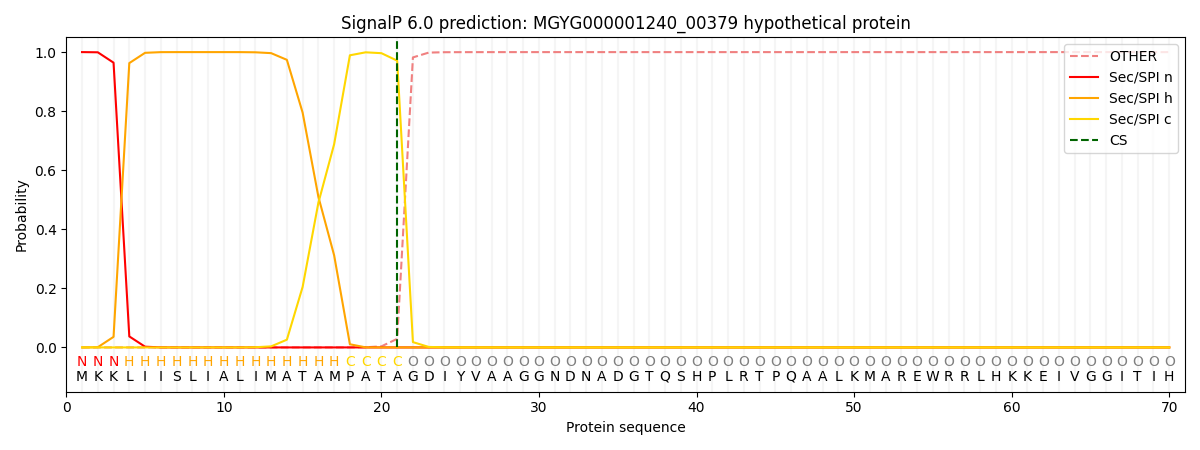
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000303 | 0.998946 | 0.000194 | 0.000182 | 0.000175 | 0.000167 |