You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001240_00772
You are here: Home > Sequence: MGYG000001240_00772
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900557405 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900557405 | |||||||||||
| CAZyme ID | MGYG000001240_00772 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 43115; End: 44635 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 198 | 474 | 2e-43 | 0.9340277777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02773 | PLN02773 | 6.14e-14 | 200 | 402 | 9 | 213 | pectinesterase |
| PLN02682 | PLN02682 | 5.99e-10 | 188 | 398 | 59 | 270 | pectinesterase family protein |
| PLN02176 | PLN02176 | 1.71e-09 | 178 | 402 | 26 | 247 | putative pectinesterase |
| PLN02745 | PLN02745 | 2.32e-08 | 187 | 392 | 276 | 462 | Putative pectinesterase/pectinesterase inhibitor |
| PLN02634 | PLN02634 | 3.64e-08 | 198 | 375 | 58 | 234 | probable pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT65269.1 | 4.67e-316 | 2 | 495 | 37 | 530 |
| QVJ81037.1 | 1.11e-222 | 4 | 494 | 3 | 497 |
| ADE82670.1 | 7.53e-220 | 4 | 494 | 19 | 513 |
| QBI04917.1 | 9.69e-149 | 25 | 494 | 9 | 485 |
| QJD94164.1 | 2.14e-148 | 14 | 494 | 6 | 486 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FHN5 | 1.25e-06 | 178 | 353 | 202 | 387 | Probable pectinesterase/pectinesterase inhibitor 59 OS=Arabidopsis thaliana OX=3702 GN=PME59 PE=2 SV=1 |
| Q9LVQ0 | 1.29e-06 | 198 | 354 | 7 | 161 | Pectinesterase 31 OS=Arabidopsis thaliana OX=3702 GN=PME31 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
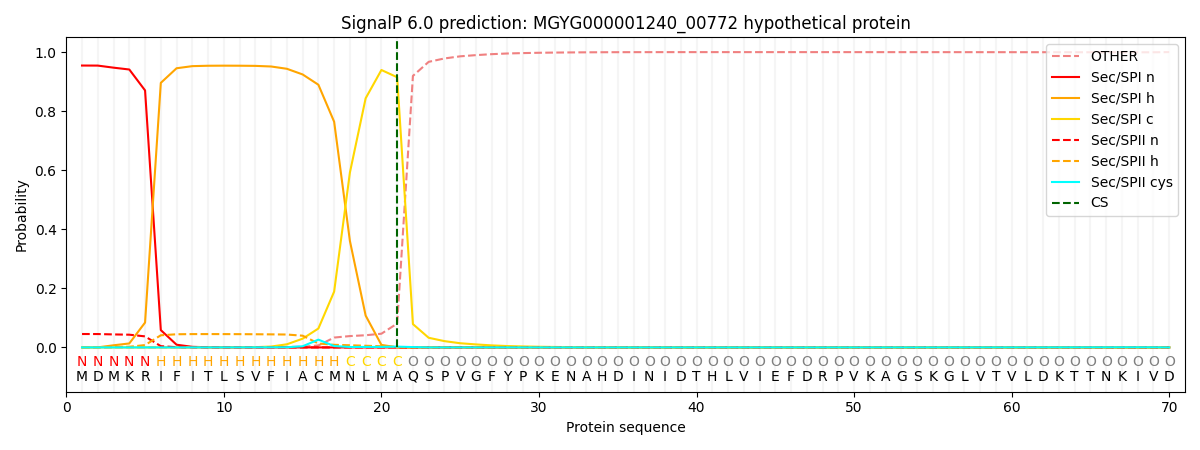
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000847 | 0.948549 | 0.049804 | 0.000261 | 0.000250 | 0.000248 |
