You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001254_02526
You are here: Home > Sequence: MGYG000001254_02526
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
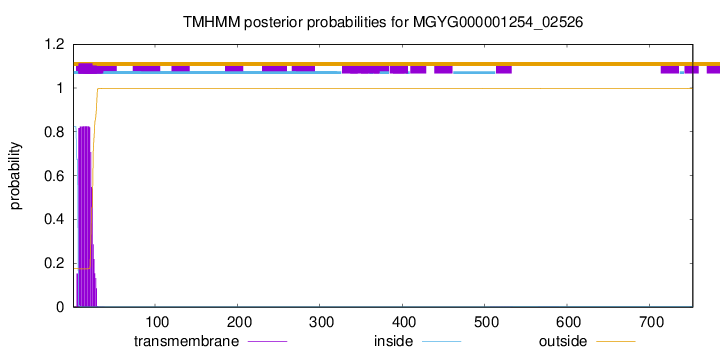
TMHMM annotations
Basic Information help
| Species | TF01-11 sp900759665 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp900759665 | |||||||||||
| CAZyme ID | MGYG000001254_02526 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10008; End: 12269 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.57e-21 | 377 | 639 | 22 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 5.47e-04 | 43 | 104 | 1 | 54 | Bacterial SH3 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHQ61172.1 | 1.37e-156 | 31 | 745 | 35 | 760 |
| BCN29841.1 | 4.00e-151 | 1 | 751 | 1 | 888 |
| BCJ93456.1 | 2.18e-144 | 2 | 751 | 4 | 894 |
| BCJ97942.1 | 3.21e-140 | 3 | 751 | 2 | 906 |
| CUH91961.1 | 1.81e-134 | 41 | 751 | 145 | 911 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PI8_A | 1.01e-08 | 482 | 634 | 85 | 226 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 1.35e-08 | 482 | 634 | 85 | 226 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 4.43e-06 | 411 | 639 | 37 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 1.99e-10 | 411 | 639 | 679 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q8CPQ1 | 2.74e-07 | 411 | 639 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 6.20e-07 | 411 | 639 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 6.20e-07 | 411 | 639 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
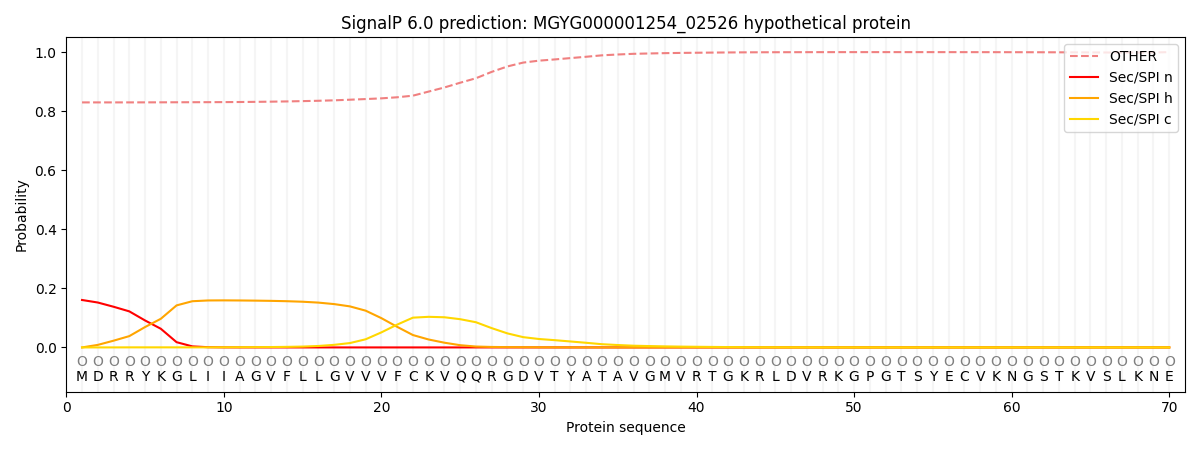
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.837220 | 0.151578 | 0.010172 | 0.000329 | 0.000211 | 0.000500 |