You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001260_01208
You are here: Home > Sequence: MGYG000001260_01208
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acinetobacter johnsonii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Moraxellaceae; Acinetobacter; Acinetobacter johnsonii | |||||||||||
| CAZyme ID | MGYG000001260_01208 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase F | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1141; End: 2253 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 222 | 363 | 2.5e-24 | 0.8296296296296296 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13403 | MLTF-like | 2.20e-67 | 213 | 363 | 1 | 161 | membrane-bound lytic murein transglycosylase F (MLTF) and similar proteins. This subfamily includes membrane-bound lytic murein transglycosylase F (MltF, murein lyase F) that degrades murein glycan strands. It is responsible for catalyzing the release of 1,6-anhydromuropeptides from peptidoglycan. Lytic transglycosylase catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do goose-type lysozymes. However, in addition, it also makes a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| PRK10859 | PRK10859 | 1.37e-65 | 9 | 357 | 2 | 446 | membrane-bound lytic murein transglycosylase MltF. |
| COG4623 | MltF | 4.06e-52 | 86 | 361 | 158 | 431 | Membrane-bound lytic murein transglycosylase MltF [Cell wall/membrane/envelope biogenesis, Signal transduction mechanisms]. |
| pfam01464 | SLT | 4.39e-24 | 217 | 323 | 5 | 112 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd00254 | LT-like | 1.05e-22 | 226 | 359 | 3 | 111 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALV72230.1 | 1.04e-269 | 1 | 370 | 1 | 370 |
| QBK70507.1 | 1.04e-269 | 1 | 370 | 1 | 370 |
| QPF34229.1 | 1.48e-269 | 1 | 370 | 1 | 370 |
| QEK35121.1 | 1.48e-269 | 1 | 370 | 1 | 370 |
| QKY91350.1 | 4.24e-269 | 1 | 370 | 1 | 370 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OZ9_A | 6.67e-42 | 158 | 330 | 206 | 381 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with isoleucine [Pseudomonas aeruginosa PAO1] |
| 4OYV_A | 7.59e-42 | 158 | 330 | 213 | 388 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with leucine [Pseudomonas aeruginosa PAO1] |
| 4OWD_A | 7.59e-42 | 158 | 330 | 213 | 388 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with cysteine [Pseudomonas aeruginosa PAO1],4OXV_A Crystal structure of MltF from Pseudomonas aeruginosa complexed with valine [Pseudomonas aeruginosa PADK2_CF510],4P0G_A Crystal structure of MltF from Pseudomonas aeruginosa complexed with bulgecin and muropeptide [Pseudomonas aeruginosa PAO1],4P11_A Native crystal structure of MltF Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5AA4_A | 1.40e-41 | 158 | 330 | 199 | 374 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA4_C Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013] |
| 5AA4_B | 1.40e-41 | 158 | 330 | 199 | 374 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA4_D Crystal structure of MltF from Pseudomonas aeruginosa in complex with cell-wall tetrapeptide [Pseudomonas aeruginosa BWHPSA013] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A4VP14 | 9.92e-43 | 145 | 330 | 222 | 411 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas stutzeri (strain A1501) OX=379731 GN=mltF PE=3 SV=2 |
| Q02RN8 | 3.88e-42 | 150 | 330 | 229 | 412 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas aeruginosa (strain UCBPP-PA14) OX=208963 GN=mltF PE=3 SV=2 |
| A1U3J1 | 4.76e-42 | 148 | 334 | 222 | 412 | Membrane-bound lytic murein transglycosylase F OS=Marinobacter nauticus (strain ATCC 700491 / DSM 11845 / VT8) OX=351348 GN=mltF PE=3 SV=1 |
| Q9HXN1 | 1.05e-40 | 158 | 330 | 238 | 413 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=mltF PE=1 SV=2 |
| A6V102 | 1.05e-40 | 158 | 330 | 238 | 413 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas aeruginosa (strain PA7) OX=381754 GN=mltF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
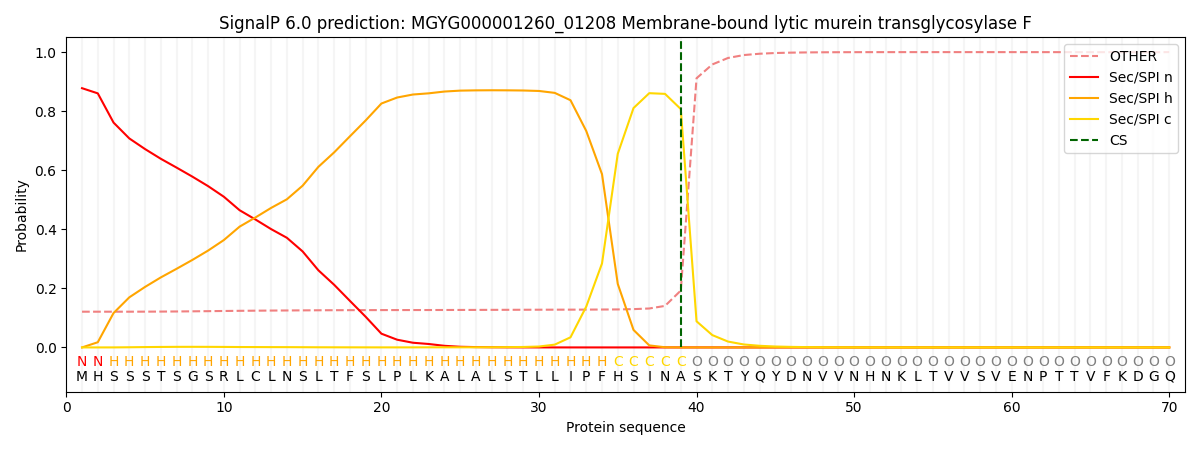
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.124801 | 0.872655 | 0.001454 | 0.000559 | 0.000254 | 0.000239 |
