You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001261_00597
You are here: Home > Sequence: MGYG000001261_00597
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caulobacter sp903900155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; Caulobacter sp903900155 | |||||||||||
| CAZyme ID | MGYG000001261_00597 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5832; End: 7307 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 181 | 446 | 1.8e-39 | 0.8888888888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02488 | PLN02488 | 1.08e-13 | 181 | 472 | 199 | 485 | probable pectinesterase/pectinesterase inhibitor |
| PLN02176 | PLN02176 | 4.48e-12 | 181 | 466 | 41 | 315 | putative pectinesterase |
| COG4677 | PemB | 1.17e-11 | 158 | 363 | 64 | 317 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 5.32e-10 | 181 | 342 | 2 | 170 | Pectinesterase. |
| PLN02773 | PLN02773 | 1.77e-09 | 178 | 406 | 3 | 279 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL01965.1 | 9.10e-227 | 21 | 491 | 42 | 508 |
| QJD94164.1 | 7.08e-112 | 1 | 488 | 1 | 491 |
| QBE65504.1 | 3.59e-108 | 10 | 490 | 15 | 509 |
| QBI04917.1 | 9.08e-107 | 20 | 490 | 8 | 492 |
| QJE01968.1 | 3.86e-104 | 24 | 490 | 32 | 510 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3EAY9 | 5.53e-09 | 164 | 303 | 170 | 317 | Probable pectinesterase 30 OS=Arabidopsis thaliana OX=3702 GN=PME30 PE=2 SV=1 |
| Q1PEC0 | 7.65e-09 | 181 | 303 | 216 | 344 | Probable pectinesterase/pectinesterase inhibitor 42 OS=Arabidopsis thaliana OX=3702 GN=PME42 PE=2 SV=1 |
| Q84JX1 | 7.65e-09 | 181 | 303 | 216 | 344 | Probable pectinesterase/pectinesterase inhibitor 19 OS=Arabidopsis thaliana OX=3702 GN=PME19 PE=2 SV=1 |
| Q4PT34 | 1.83e-07 | 171 | 307 | 34 | 178 | Probable pectinesterase 56 OS=Arabidopsis thaliana OX=3702 GN=PME56 PE=2 SV=1 |
| Q9LSP1 | 5.92e-07 | 140 | 275 | 4 | 140 | Probable pectinesterase 67 OS=Arabidopsis thaliana OX=3702 GN=PME67 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
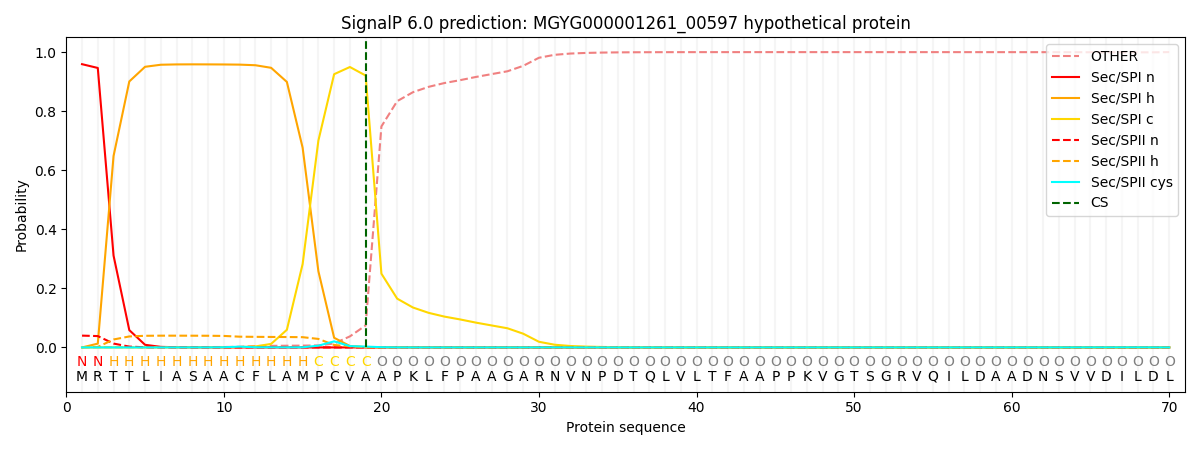
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002175 | 0.954475 | 0.042412 | 0.000439 | 0.000254 | 0.000210 |
