You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001261_02182
You are here: Home > Sequence: MGYG000001261_02182
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caulobacter sp903900155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; Caulobacter sp903900155 | |||||||||||
| CAZyme ID | MGYG000001261_02182 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8983; End: 11037 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 2 | 681 | 1.2e-209 | 0.8507281553398058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 0.0 | 2 | 488 | 378 | 873 | alpha-L-rhamnosidase. |
| PRK10150 | PRK10150 | 5.69e-05 | 530 | 638 | 14 | 136 | beta-D-glucuronidase; Provisional |
| PRK14963 | PRK14963 | 4.45e-04 | 328 | 501 | 213 | 378 | DNA polymerase III subunits gamma and tau; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AHE55710.1 | 8.88e-189 | 1 | 682 | 373 | 1073 |
| AOH83238.1 | 1.78e-188 | 1 | 680 | 425 | 1136 |
| QXD15381.1 | 5.02e-186 | 1 | 680 | 411 | 1124 |
| AIT08070.1 | 1.83e-185 | 1 | 680 | 410 | 1121 |
| QOY84991.1 | 7.15e-184 | 1 | 680 | 425 | 1120 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q2F_A | 1.70e-152 | 1 | 680 | 440 | 1138 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
| 5MQM_A | 1.33e-87 | 1 | 683 | 397 | 1101 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 3.49e-87 | 1 | 683 | 397 | 1101 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 1.87e-86 | 1 | 682 | 240 | 1157 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
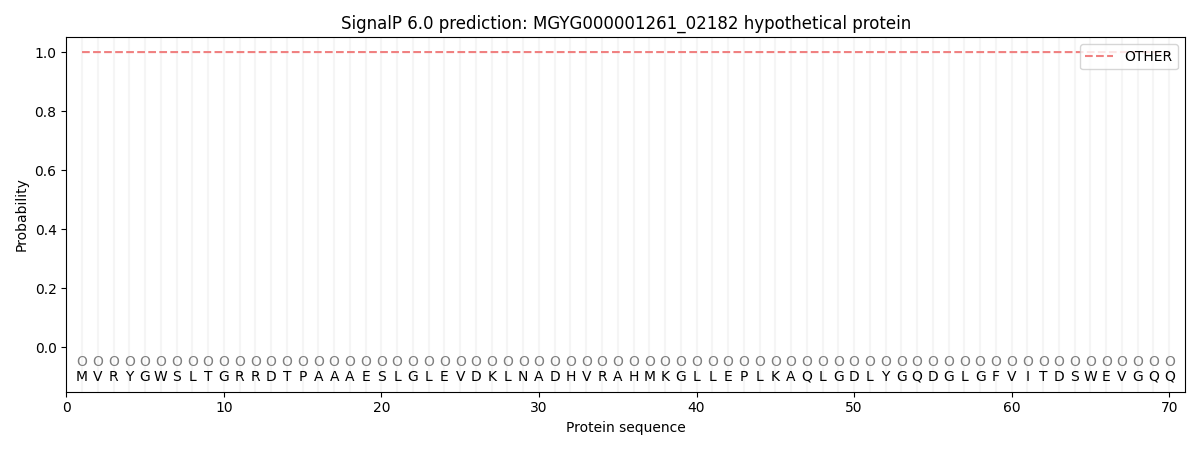
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000079 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
