You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001261_03413
You are here: Home > Sequence: MGYG000001261_03413
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caulobacter sp903900155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; Caulobacter sp903900155 | |||||||||||
| CAZyme ID | MGYG000001261_03413 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1877; End: 3232 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN02432 | PLN02432 | 4.93e-14 | 37 | 264 | 13 | 212 | putative pectinesterase |
| PLN02488 | PLN02488 | 1.33e-12 | 34 | 230 | 196 | 368 | probable pectinesterase/pectinesterase inhibitor |
| PLN02497 | PLN02497 | 2.99e-11 | 37 | 267 | 34 | 249 | probable pectinesterase |
| PLN02682 | PLN02682 | 8.33e-11 | 37 | 264 | 71 | 287 | pectinesterase family protein |
| pfam01095 | Pectinesterase | 1.02e-10 | 36 | 264 | 1 | 218 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACB77364.1 | 4.86e-150 | 23 | 447 | 23 | 452 |
| AFN75583.1 | 9.52e-87 | 36 | 416 | 23 | 371 |
| QDV72704.1 | 7.58e-79 | 49 | 414 | 35 | 369 |
| QDV77276.1 | 7.58e-79 | 49 | 414 | 35 | 369 |
| QEG36438.1 | 1.16e-77 | 36 | 435 | 43 | 412 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 2.98e-09 | 47 | 228 | 18 | 194 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3EAY9 | 3.68e-10 | 34 | 267 | 186 | 406 | Probable pectinesterase 30 OS=Arabidopsis thaliana OX=3702 GN=PME30 PE=2 SV=1 |
| Q84JX1 | 3.86e-10 | 34 | 267 | 213 | 433 | Probable pectinesterase/pectinesterase inhibitor 19 OS=Arabidopsis thaliana OX=3702 GN=PME19 PE=2 SV=1 |
| Q1PEC0 | 2.78e-09 | 34 | 267 | 213 | 433 | Probable pectinesterase/pectinesterase inhibitor 42 OS=Arabidopsis thaliana OX=3702 GN=PME42 PE=2 SV=1 |
| Q9SIJ9 | 4.05e-08 | 25 | 264 | 39 | 252 | Putative pectinesterase 11 OS=Arabidopsis thaliana OX=3702 GN=PME11 PE=3 SV=1 |
| Q43043 | 5.87e-08 | 35 | 264 | 57 | 275 | Pectinesterase OS=Petunia integrifolia OX=4103 GN=PPE1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
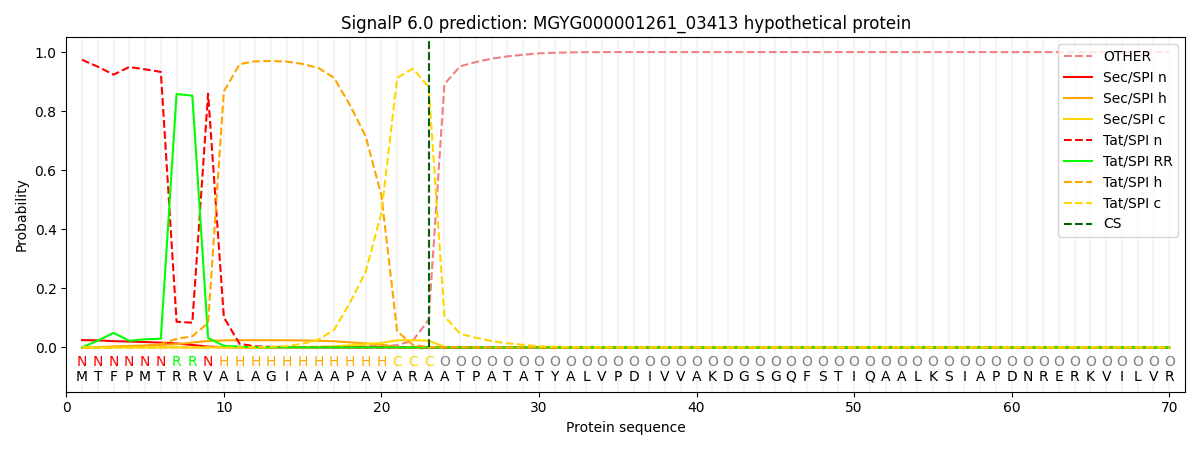
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000813 | 0.024091 | 0.000044 | 0.974113 | 0.000908 | 0.000027 |
