You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001261_03468
You are here: Home > Sequence: MGYG000001261_03468
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Caulobacter sp903900155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Caulobacter; Caulobacter sp903900155 | |||||||||||
| CAZyme ID | MGYG000001261_03468 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3; End: 1073 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 41 | 350 | 2.5e-80 | 0.9603960396039604 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 5.29e-104 | 66 | 351 | 58 | 340 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 5.30e-86 | 43 | 350 | 11 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 5.18e-85 | 75 | 344 | 1 | 259 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS20276.1 | 6.88e-150 | 4 | 351 | 3 | 354 |
| AZH14072.1 | 1.85e-141 | 4 | 351 | 3 | 349 |
| ATC25929.1 | 1.85e-141 | 4 | 351 | 3 | 349 |
| ACL96602.1 | 1.50e-140 | 4 | 351 | 3 | 349 |
| ATC29873.1 | 1.50e-140 | 4 | 351 | 3 | 349 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6WQW_A | 4.26e-54 | 42 | 353 | 14 | 331 | ChainA, Beta-xylanase [Thermobacillus composti] |
| 2Q8X_A | 3.58e-51 | 61 | 342 | 33 | 322 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 1N82_A | 3.58e-51 | 61 | 342 | 33 | 322 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 7NL2_A | 1.69e-50 | 39 | 337 | 17 | 326 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 3MUI_A | 2.76e-50 | 61 | 342 | 33 | 322 | Enzyme-Substrateinteractions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUI_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P45703 | 1.83e-46 | 42 | 342 | 16 | 321 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=1 SV=1 |
| O69231 | 1.04e-45 | 42 | 336 | 16 | 316 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| O69230 | 1.95e-43 | 42 | 350 | 379 | 709 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| P40944 | 1.06e-42 | 42 | 350 | 364 | 677 | Endo-1,4-beta-xylanase A OS=Caldicellulosiruptor sp. (strain Rt8B.4) OX=28238 GN=xynA PE=3 SV=1 |
| Q59675 | 3.52e-42 | 36 | 350 | 251 | 595 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
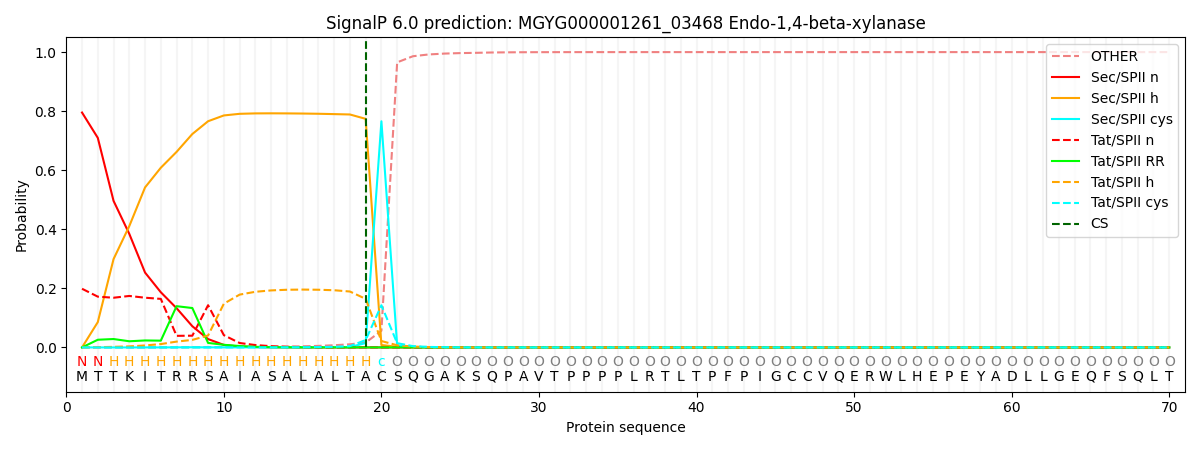
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000165 | 0.000668 | 0.795381 | 0.005137 | 0.198651 | 0.000006 |
