You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001263_00171
You are here: Home > Sequence: MGYG000001263_00171
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acidovorax sp000302535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Acidovorax; Acidovorax sp000302535 | |||||||||||
| CAZyme ID | MGYG000001263_00171 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64449; End: 66140 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 15 | 510 | 4.1e-88 | 0.9111111111111111 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 3.13e-43 | 9 | 524 | 3 | 505 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 1.21e-17 | 31 | 384 | 23 | 375 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231 | PMT_2 | 2.47e-13 | 69 | 232 | 1 | 159 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1287 | Stt3 | 0.001 | 67 | 370 | 84 | 379 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIL19303.1 | 2.53e-194 | 13 | 559 | 5 | 549 |
| QDA58490.1 | 2.27e-192 | 11 | 559 | 3 | 549 |
| QNN71169.1 | 3.10e-190 | 13 | 559 | 5 | 550 |
| ATE73848.1 | 6.79e-186 | 5 | 561 | 3 | 576 |
| ALN88166.1 | 4.69e-185 | 18 | 561 | 1 | 561 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 3.55e-20 | 14 | 340 | 32 | 356 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q48HY9 | 1.64e-23 | 15 | 350 | 7 | 333 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas savastanoi pv. phaseolicola (strain 1448A / Race 6) OX=264730 GN=arnT PE=3 SV=1 |
| Q4ZSZ0 | 1.02e-22 | 13 | 354 | 7 | 338 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
| Q2NRV9 | 7.64e-21 | 31 | 386 | 22 | 373 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Sodalis glossinidius (strain morsitans) OX=343509 GN=arnT2 PE=3 SV=1 |
| Q6D2F3 | 1.36e-19 | 19 | 380 | 8 | 373 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 1.79e-19 | 37 | 469 | 28 | 451 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
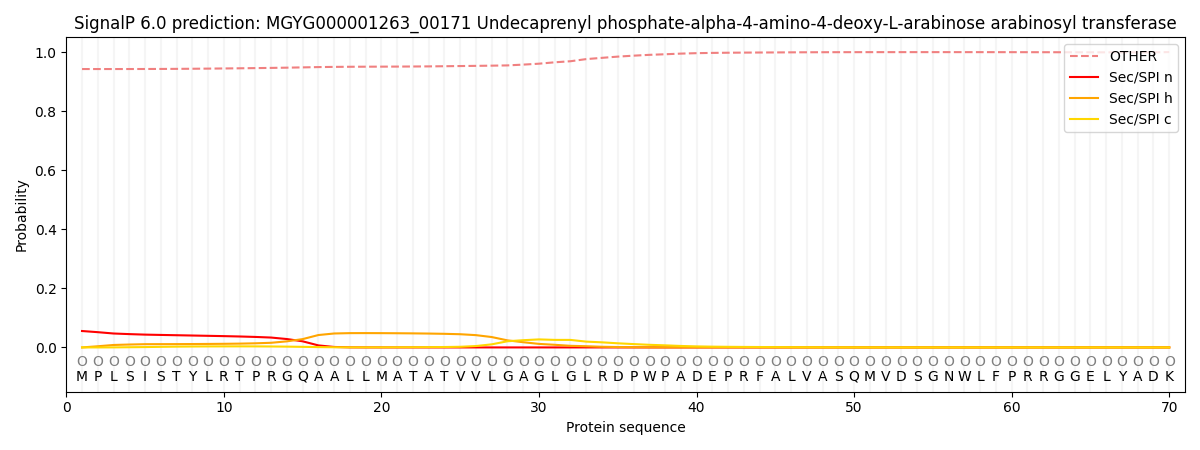
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.946140 | 0.051118 | 0.001054 | 0.000184 | 0.000120 | 0.001397 |
TMHMM Annotations download full data without filtering help
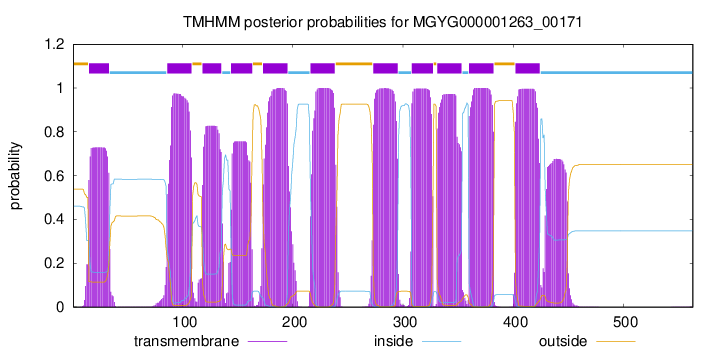
| start | end |
|---|---|
| 15 | 33 |
| 86 | 108 |
| 118 | 135 |
| 144 | 163 |
| 173 | 195 |
| 216 | 238 |
| 273 | 295 |
| 308 | 327 |
| 331 | 353 |
| 360 | 382 |
| 402 | 424 |
