You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001264_00806
You are here: Home > Sequence: MGYG000001264_00806
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acidovorax temperans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Acidovorax; Acidovorax temperans | |||||||||||
| CAZyme ID | MGYG000001264_00806 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11487; End: 12809 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03025 | EPS_sugtrans | 6.16e-89 | 28 | 440 | 4 | 445 | exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase. Members of this family are generally found near other genes involved in the biosynthesis of a variety of exopolysaccharides. These proteins consist of two fused domains, an N-terminal hydrophobic domain of generally low conservation and a highly conserved C-terminal sugar transferase domain (pfam02397). Characterized and partially characterized members of this subfamily include Salmonella WbaP (originally RfbP), E. coli WcaJ, Methylobacillus EpsB, Xanthomonas GumD, Vibrio CpsA, Erwinia AmsG, Group B Streptococcus CpsE (originally CpsD), and Streptococcus suis Cps2E. Each of these is believed to act in transferring the sugar from, for instance, UDP-glucose or UDP-galactose, to a lipid carrier such as undecaprenyl phosphate as the first (priming) step in the synthesis of an oligosaccharide "block". This function is encoded in the C-terminal domain. The liposaccharide is believed to be subsequently transferred through a "flippase" function from the cytoplasmic to the periplasmic face of the inner membrane by the N-terminal domain. Certain closely related transferase enzymes, such as Sinorhizobium ExoY and Lactococcus EpsD, lack the N-terminal domain and are not found by this model. |
| pfam02397 | Bac_transf | 4.77e-75 | 273 | 435 | 21 | 181 | Bacterial sugar transferase. This Pfam family represents a conserved region from a number of different bacterial sugar transferases, involved in diverse biosynthesis pathways. |
| TIGR03023 | WcaJ_sugtrans | 5.43e-67 | 252 | 436 | 259 | 446 | Undecaprenyl-phosphate glucose phosphotransferase. This family of proteins encompasses the E. coli WcaJ protein involved in colanic acid biosynthesis, the Methylobacillus EpsB protein involved in methanolan biosynthesis, as well as the GumD protein involved in the biosynthesis of xanthan. All of these are closely related to the well-characterized WbaP (formerly RfbP) protein, which is the first enzyme in O-antigen biosynthesis in Salmonella typhimurium. The enzyme transfers galactose from UDP-galactose (NOTE: not glucose) to a polyprenyl carrier (utilizing the highly conserved C-terminal sugar transferase domain, pfam02397) a reaction which takes place at the cytoplasmic face of the inner membrane. The N-terminal hydrophobic domain is then believed to facilitate the "flippase" function of transferring the liposaccharide unit from the cytoplasmic face to the periplasmic face of the inner membrane. Most of these genes are found within large operons dedicated to the production of complex exopolysaccharides such as the enterobacterial O-antigen. Colanic acid biosynthesis utilizes a glucose-undecaprenyl carrier, knockout of EpsB abolishes incorporation of UDP-glucose into the lipid phase, and the C-terminal portion of GumD has been shown to be responsible for the glucosyl-1-transferase activity. |
| COG2148 | WcaJ | 1.08e-66 | 239 | 439 | 27 | 225 | Sugar transferase involved in LPS biosynthesis (colanic, teichoic acid) [Cell wall/membrane/envelope biogenesis]. |
| TIGR03013 | EpsB_2 | 6.18e-65 | 251 | 438 | 254 | 442 | sugar transferase, PEP-CTERM system associated. Members of this protein family belong to the family of bacterial sugar transferases (pfam02397). Nearly all are found in species that encode the PEP-CTERM/exosortase system predicted to act in protein sorting in a number of Gram-negative bacteria (notable exceptions appear to include Magnetococcus sp. MC-1 and Myxococcus xanthus DK 1622 ). These genes are generally found near one or more of the PrsK, PrsR or PrsT genes that have been related to the PEP-CTERM system by phylogenetic profiling methods. The nature of the sugar transferase reaction catalyzed by members of this clade is unknown and may conceivably be variable with respect to substrate by species. These proteins are homologs of the EpsB protein found in Methylobacillus sp. strain 12S, which is also associated with a PEP-CTERM system, but of a distinct type. A name which appears attached to a number of genes (by transitive annotation) in this family is "undecaprenyl-phosphate galactose phosphotransferase", which comes from relatively distant characterized enterobacterial homologs, and is considerably more specific than warranted from the currently available evidence. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALK88913.1 | 2.16e-130 | 9 | 440 | 380 | 817 |
| AWW69811.1 | 6.84e-64 | 190 | 440 | 275 | 527 |
| ATD00364.1 | 1.35e-35 | 253 | 438 | 399 | 596 |
| BBN81660.1 | 1.44e-33 | 253 | 438 | 398 | 595 |
| AIY66659.1 | 3.18e-32 | 253 | 438 | 399 | 596 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5W7L_A | 5.05e-28 | 252 | 429 | 10 | 180 | Structureof Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826],5W7L_B Structure of Campylobacter concisus PglC I57M/Q175M variant [Campylobacter concisus 13826] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B3FN88 | 5.83e-70 | 203 | 440 | 185 | 423 | UDP-N-acetylgalactosamine-undecaprenyl-phosphate N-acetylgalactosaminephosphotransferase OS=Aeromonas hydrophila OX=644 GN=wecA PE=1 SV=2 |
| Q9ABR0 | 1.95e-36 | 248 | 435 | 72 | 262 | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase OS=Caulobacter vibrioides (strain ATCC 19089 / CB15) OX=190650 GN=pssY PE=1 SV=1 |
| P71241 | 1.78e-34 | 247 | 437 | 260 | 458 | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase OS=Escherichia coli (strain K12) OX=83333 GN=wcaJ PE=1 SV=2 |
| Q56770 | 4.41e-34 | 243 | 430 | 283 | 474 | UDP-glucose:undecaprenyl-phosphate glucose-1-phosphate transferase OS=Xanthomonas campestris pv. campestris OX=340 GN=gumD PE=1 SV=1 |
| P10498 | 9.43e-34 | 252 | 429 | 5 | 189 | Exopolysaccharide production protein PSS OS=Rhizobium leguminosarum bv. phaseoli OX=385 GN=pss PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
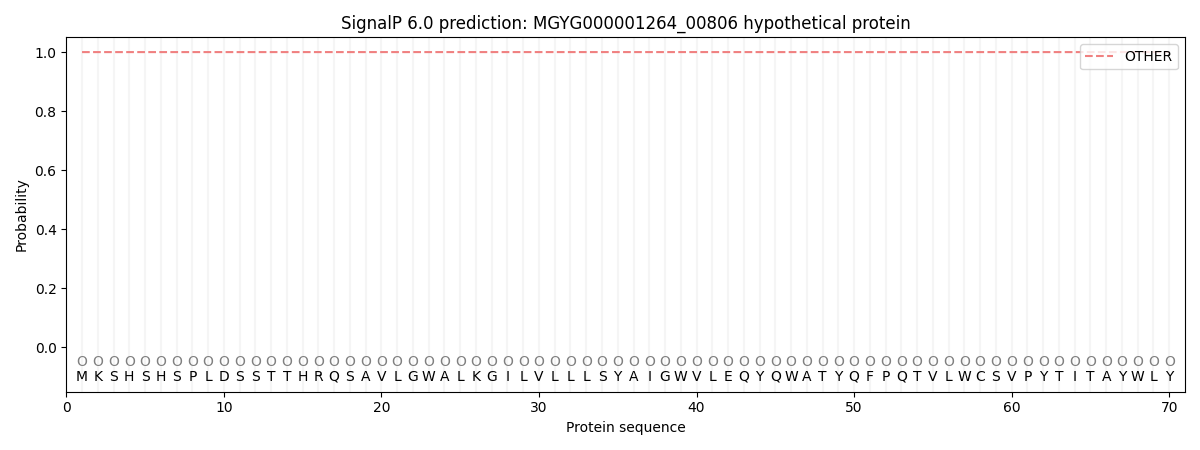
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000029 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help

| start | end |
|---|---|
| 19 | 41 |
| 51 | 70 |
| 83 | 102 |
| 106 | 125 |
| 258 | 279 |
