You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001265_00116
You are here: Home > Sequence: MGYG000001265_00116
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
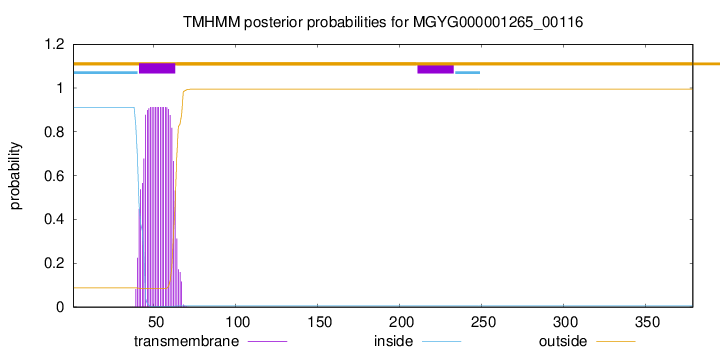
TMHMM annotations
Basic Information help
| Species | Methylobacterium sp002778835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Beijerinckiaceae; Methylobacterium; Methylobacterium sp002778835 | |||||||||||
| CAZyme ID | MGYG000001265_00116 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 91117; End: 92256 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01464 | SLT | 0.005 | 163 | 208 | 1 | 45 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWN42370.1 | 2.96e-194 | 1 | 379 | 1 | 383 |
| ACB27551.1 | 4.08e-186 | 1 | 378 | 1 | 383 |
| AWN37034.1 | 3.02e-185 | 1 | 378 | 1 | 391 |
| QGY00938.1 | 9.04e-183 | 1 | 378 | 1 | 383 |
| APT34194.1 | 4.25e-181 | 1 | 376 | 1 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3BKH_A | 1.23e-17 | 176 | 328 | 115 | 243 | ChainA, lytic transglycosylase [Pseudomonas phage phiKZ],3BKV_A Chain A, lytic transglycosylase [Pseudomonas phage phiKZ] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8SCY1 | 9.69e-17 | 157 | 328 | 1878 | 2027 | Peptidoglycan hydrolase gp181 OS=Pseudomonas phage phiKZ OX=169683 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
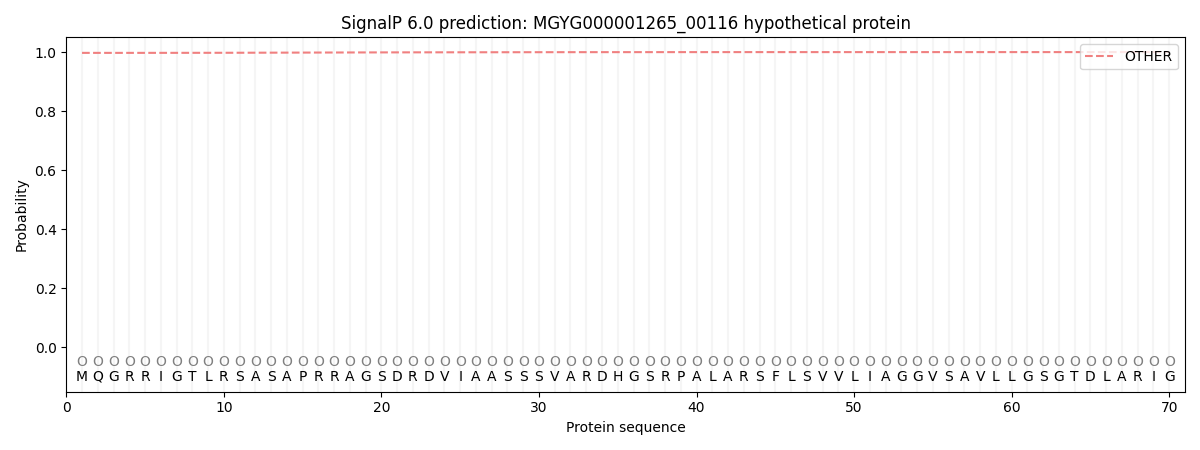
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.998441 | 0.001546 | 0.000001 | 0.000001 | 0.000000 | 0.000002 |