You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001265_03558
You are here: Home > Sequence: MGYG000001265_03558
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Methylobacterium sp002778835 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Beijerinckiaceae; Methylobacterium; Methylobacterium sp002778835 | |||||||||||
| CAZyme ID | MGYG000001265_03558 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 111; End: 1835 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 28 | 506 | 4.8e-97 | 0.8814814814814815 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 3.69e-41 | 44 | 516 | 23 | 492 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279 | arnT | 1.67e-05 | 50 | 452 | 30 | 428 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTI80784.1 | 3.25e-99 | 34 | 565 | 18 | 548 |
| QYC12562.1 | 3.11e-78 | 36 | 574 | 21 | 561 |
| QYC09774.1 | 3.11e-78 | 36 | 574 | 21 | 561 |
| AXE66036.1 | 1.06e-55 | 44 | 388 | 27 | 381 |
| ABI78434.1 | 7.16e-54 | 44 | 388 | 27 | 381 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5EZM_A | 2.84e-10 | 55 | 387 | 58 | 383 | CrystalStructure of ArnT from Cupriavidus metallidurans in the apo state [Cupriavidus metallidurans CH34],5F15_A Crystal Structure of ArnT from Cupriavidus metallidurans bound to Undecaprenyl phosphate [Cupriavidus metallidurans CH34] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3KCB9 | 8.56e-10 | 55 | 376 | 31 | 341 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT2 PE=3 SV=1 |
| B2VBI7 | 1.84e-08 | 55 | 361 | 31 | 327 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=arnT PE=3 SV=1 |
| Q4ZSZ0 | 2.42e-08 | 55 | 361 | 33 | 327 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Pseudomonas syringae pv. syringae (strain B728a) OX=205918 GN=arnT PE=3 SV=1 |
| C5BDQ8 | 2.43e-08 | 43 | 361 | 22 | 327 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
| Q4K884 | 3.33e-08 | 55 | 379 | 64 | 377 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 2 OS=Pseudomonas fluorescens (strain ATCC BAA-477 / NRRL B-23932 / Pf-5) OX=220664 GN=arnT2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.959489 | 0.040273 | 0.000097 | 0.000031 | 0.000029 | 0.000084 |
TMHMM Annotations download full data without filtering help
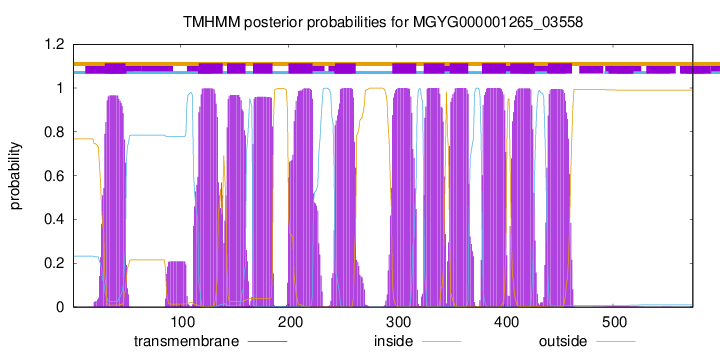
| start | end |
|---|---|
| 30 | 49 |
| 117 | 139 |
| 143 | 160 |
| 167 | 185 |
| 200 | 222 |
| 243 | 262 |
| 296 | 318 |
| 325 | 344 |
| 349 | 366 |
| 379 | 401 |
| 405 | 427 |
| 440 | 462 |
