You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001270_00186
You are here: Home > Sequence: MGYG000001270_00186
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium sp902373425 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium sp902373425 | |||||||||||
| CAZyme ID | MGYG000001270_00186 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3797; End: 7567 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13402 | LT_TF-like | 6.35e-45 | 984 | 1102 | 1 | 110 | lytic transglycosylase-like domain of tail fiber-like proteins and similar domains. These tail fiber-like proteins are multi-domain proteins that include a lytic transglycosylase (LT) domain. Members of the LT family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, and the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| COG3953 | SLT | 1.81e-21 | 978 | 1102 | 9 | 133 | SLT domain protein [Mobilome: prophages, transposons]. |
| COG5412 | COG5412 | 3.46e-09 | 390 | 765 | 88 | 492 | Phage-related protein [Mobilome: prophages, transposons]. |
| pfam01464 | SLT | 3.51e-08 | 991 | 1050 | 19 | 73 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| cd00254 | LT-like | 9.73e-08 | 991 | 1067 | 8 | 69 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRP14781.1 | 0.0 | 1 | 1256 | 1 | 1256 |
| QRP10329.1 | 0.0 | 1 | 1256 | 1 | 1256 |
| CAB93921.2 | 2.55e-67 | 729 | 1173 | 839 | 1438 |
| QLJ05716.1 | 1.19e-57 | 738 | 1106 | 932 | 1290 |
| QLJ01544.1 | 1.95e-56 | 738 | 1106 | 947 | 1305 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P54334 | 3.15e-17 | 958 | 1106 | 1089 | 1232 | Phage-like element PBSX protein XkdO OS=Bacillus subtilis (strain 168) OX=224308 GN=xkdO PE=4 SV=2 |
| P45931 | 1.95e-15 | 958 | 1106 | 1341 | 1484 | Uncharacterized protein YqbO OS=Bacillus subtilis (strain 168) OX=224308 GN=yqbO PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999922 | 0.000101 | 0.000003 | 0.000001 | 0.000000 | 0.000001 |
TMHMM Annotations download full data without filtering help
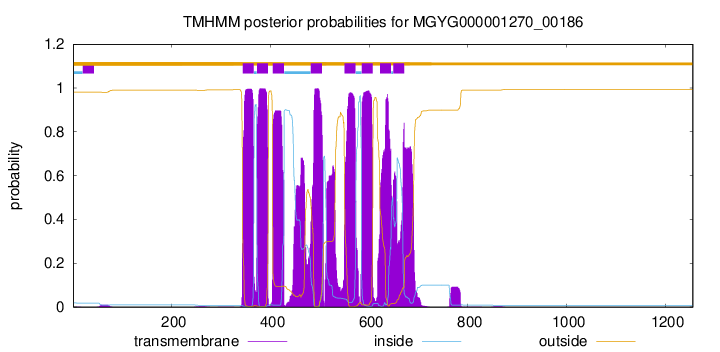
| start | end |
|---|---|
| 344 | 366 |
| 373 | 395 |
| 405 | 427 |
| 482 | 504 |
| 550 | 572 |
| 585 | 607 |
| 622 | 644 |
| 649 | 671 |
