You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001273_00001
You are here: Home > Sequence: MGYG000001273_00001
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
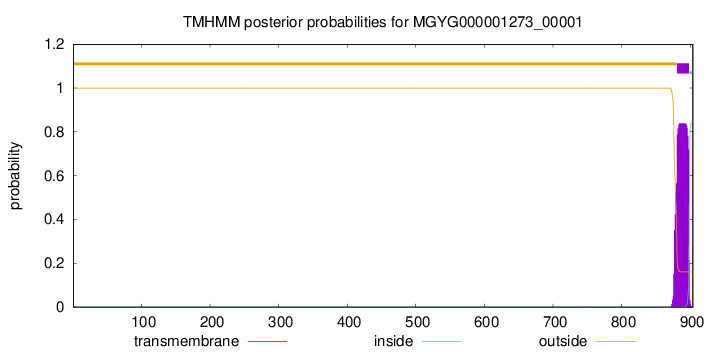
TMHMM annotations
Basic Information help
| Species | Pauljensenia sp902373435 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia sp902373435 | |||||||||||
| CAZyme ID | MGYG000001273_00001 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16; End: 2730 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 301 | 445 | 4.2e-35 | 0.9776119402985075 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 3.37e-39 | 299 | 446 | 2 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.47e-30 | 300 | 446 | 5 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam17974 | GalBD_like | 2.29e-23 | 2 | 155 | 22 | 170 | Galactose-binding domain-like. Proteins containing a galactose-binding domain-like fold can be found in several different protein families, in both eukaryotes and prokaryotes. The common function of these domains is to bind to specific ligands, such as cell-surface-attached carbohydrate substrates for galactose oxidase and sialidase, phospholipids on the outer side of the mammalian cell membrane for coagulation factor Va, membrane-anchored ephrin for the Eph family of receptor tyrosine kinases, and a complex of broken single-stranded DNA and DNA polymerase beta for XRCC1. The structure of the galactose-binding domain-like members consists of a beta-sandwich, in which the strands making up the sheets exhibit a jellyroll fold. |
| pfam10633 | NPCBM_assoc | 3.79e-14 | 196 | 274 | 1 | 78 | NPCBM-associated, NEW3 domain of alpha-galactosidase. The English-language version of the first reference can be found on pages 388-399 of the above. This domain has been named NEW3 but its actual function is not known. It is found on proteins which are bacterial galactosidases. The domain is associated with the NPCBM family, pfam08305, a novel putative carbohydrate binding module found at the N-terminus of glycosyl hydrolases. |
| pfam00754 | F5_F8_type_C | 1.51e-08 | 484 | 598 | 9 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEG53803.1 | 1.12e-287 | 1 | 797 | 857 | 1643 |
| QQM68206.1 | 1.49e-283 | 1 | 794 | 861 | 1645 |
| AVM61293.1 | 2.60e-267 | 1 | 797 | 864 | 1653 |
| QQO77976.1 | 6.96e-267 | 1 | 797 | 864 | 1653 |
| QDW61529.1 | 4.08e-101 | 5 | 485 | 903 | 1371 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5A55_A | 4.95e-22 | 3 | 179 | 916 | 1108 | Thenative structure of GH101 from Streptococcus pneumoniae TIGR4 [Streptococcus pneumoniae TIGR4] |
| 5A59_A | 4.96e-22 | 3 | 179 | 914 | 1106 | Thestructure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen [Streptococcus pneumoniae TIGR4],5A5A_A The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with PNP-T-antigen [Streptococcus pneumoniae TIGR4] |
| 5A56_A | 4.96e-22 | 3 | 179 | 914 | 1106 | Thestructure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen [Streptococcus pneumoniae TIGR4] |
| 5A57_A | 4.96e-22 | 3 | 179 | 914 | 1106 | Thestructure of GH101 from Streptococcus pneumoniae TIGR4 in complex with PUGT [Streptococcus pneumoniae TIGR4] |
| 5A58_A | 4.96e-22 | 3 | 179 | 914 | 1106 | Thestructure of GH101 D764N mutant from Streptococcus pneumoniae TIGR4 in complex with serinyl T-antigen [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A9WNA0 | 1.03e-82 | 6 | 480 | 877 | 1330 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| Q2MGH6 | 3.37e-21 | 3 | 179 | 1229 | 1421 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| Q8DR60 | 3.37e-21 | 3 | 179 | 1229 | 1421 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
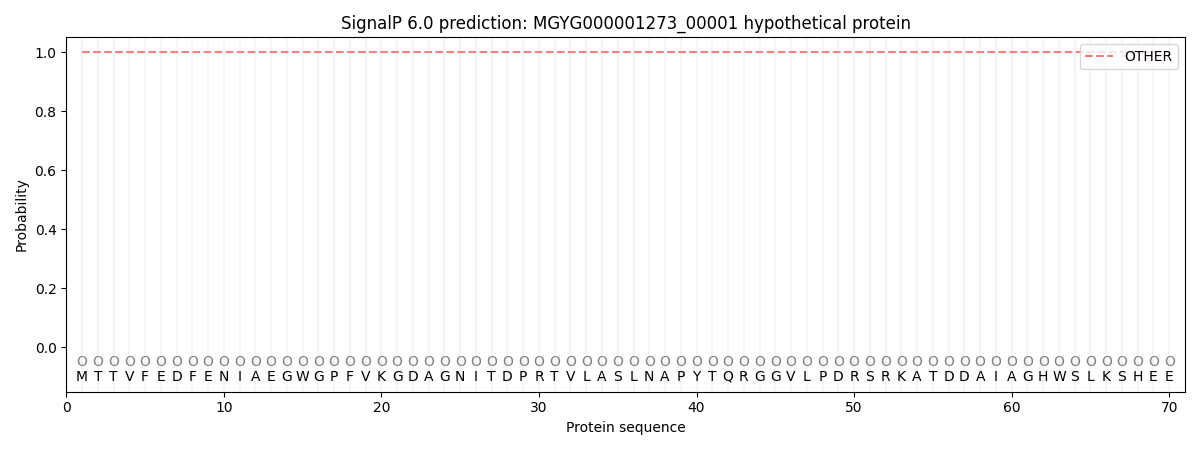
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000072 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |