You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001275_01101
You are here: Home > Sequence: MGYG000001275_01101
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-485 sp900321585 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900321585 | |||||||||||
| CAZyme ID | MGYG000001275_01101 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4568; End: 6256 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 326 | 552 | 5e-23 | 0.6732673267326733 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.23e-17 | 326 | 549 | 54 | 260 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 4.02e-09 | 320 | 559 | 117 | 344 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDO69412.1 | 3.23e-103 | 21 | 559 | 23 | 742 |
| QUT92912.1 | 3.21e-99 | 14 | 559 | 12 | 730 |
| BCS85403.1 | 1.68e-96 | 20 | 559 | 15 | 561 |
| QIH34287.1 | 5.46e-90 | 5 | 559 | 3 | 560 |
| QQT29341.1 | 4.20e-89 | 5 | 559 | 3 | 560 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1B30_A | 1.35e-10 | 330 | 556 | 105 | 302 | XYLANASEFROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH 1,2-(4-DEOXY-BETA-L-THREO-HEX-4-ENOPYRANOSYLURONIC ACID)-BETA-1,4-XYLOTRIOSE) [Penicillium simplicissimum],1B31_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, NATIVE WITH PEG200 AS CRYOPROTECTANT [Penicillium simplicissimum],1B3V_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOSE [Penicillium simplicissimum],1B3W_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOBIOSE [Penicillium simplicissimum],1B3X_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOTRIOSE [Penicillium simplicissimum],1B3Y_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOTETRAOSE [Penicillium simplicissimum],1B3Z_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM, COMPLEX WITH XYLOPENTAOSE [Penicillium simplicissimum],1BG4_A XYLANASE FROM PENICILLIUM SIMPLICISSIMUM [Penicillium simplicissimum] |
| 4XUY_A | 3.25e-10 | 330 | 556 | 105 | 302 | Crystalstructure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88],4XUY_B Crystal structure of an endo-beta-1,4-xylanase (glycoside hydrolase family 10/GH10) enzyme from Aspergillus niger [Aspergillus niger CBS 513.88] |
| 1I1W_A | 1.88e-09 | 330 | 494 | 104 | 246 | 0.89AUltra high resolution structure of a Thermostable Xylanase from Thermoascus Aurantiacus [Thermoascus aurantiacus],1I1X_A 1.11 A ATOMIC RESOLUTION STRUCTURE OF A THERMOSTABLE XYLANASE FROM THERMOASCUS AURANTIACUS [Thermoascus aurantiacus] |
| 6FHF_A | 2.01e-09 | 292 | 490 | 97 | 270 | Highlyactive enzymes by automated modular backbone assembly and sequence design [Escherichia coli] |
| 2BNJ_A | 4.48e-09 | 330 | 494 | 104 | 246 | Thexylanase TA from Thermoascus aurantiacus utilizes arabinose decorations of xylan as significant substrate specificity determinants. [Thermoascus aurantiacus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O59859 | 8.86e-11 | 330 | 556 | 130 | 327 | Endo-1,4-beta-xylanase OS=Aspergillus aculeatus OX=5053 GN=xynIA PE=3 SV=1 |
| P56588 | 7.41e-10 | 330 | 556 | 105 | 302 | Endo-1,4-beta-xylanase OS=Penicillium simplicissimum OX=69488 PE=1 SV=1 |
| Q5S7A8 | 9.02e-10 | 330 | 556 | 130 | 327 | Endo-1,4-beta-xylanase A OS=Penicillium canescens OX=5083 GN=xylA PE=1 SV=1 |
| A2QFV7 | 2.15e-09 | 330 | 556 | 130 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xlnC PE=1 SV=1 |
| C5J411 | 2.87e-09 | 330 | 556 | 130 | 327 | Probable endo-1,4-beta-xylanase C OS=Aspergillus niger OX=5061 GN=xlnC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
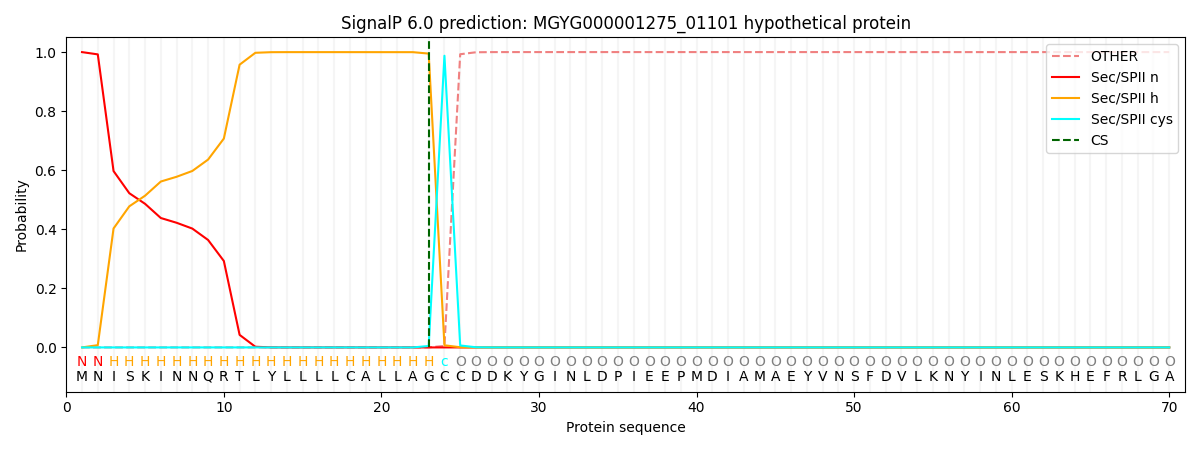
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000013 | 1.000022 | 0.000000 | 0.000000 | 0.000000 |
