You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001275_02164
Basic Information
help
| Species |
CAG-485 sp900321585
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; CAG-485 sp900321585
|
| CAZyme ID |
MGYG000001275_02164
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001275 |
2994513 |
MAG |
Italy |
Europe |
|
| Gene Location |
Start: 8126;
End: 9697
Strand: -
|
No EC number prediction in MGYG000001275_02164.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
8 |
447 |
1.5e-64 |
0.8222222222222222 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.37e-18 |
11 |
415 |
11 |
420 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| PRK13279
|
arnT |
6.98e-06 |
12 |
403 |
11 |
406 |
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| pfam13231
|
PMT_2 |
0.008 |
63 |
221 |
1 |
154 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O67270
|
2.03e-09 |
17 |
338 |
5 |
323 |
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
|
Q3KCC9
|
5.96e-07 |
21 |
325 |
37 |
339 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase 1 OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=arnT1 PE=3 SV=1 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000007
|
0.000001
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
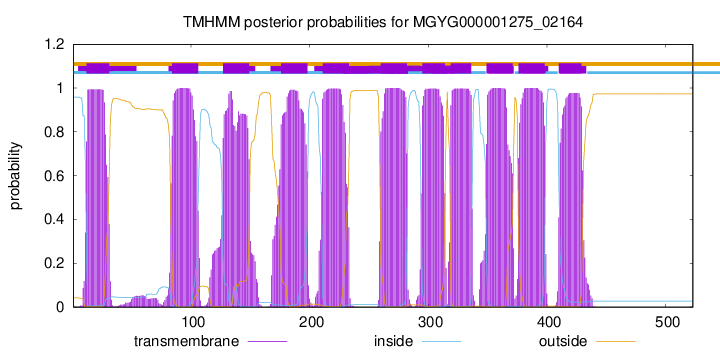
| start |
end |
| 12 |
31 |
| 84 |
106 |
| 127 |
149 |
| 176 |
198 |
| 211 |
233 |
| 260 |
282 |
| 295 |
314 |
| 319 |
336 |
| 349 |
371 |
| 376 |
398 |
| 410 |
429 |


