You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001279_00933
You are here: Home > Sequence: MGYG000001279_00933
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas_E fulva | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_E; Pseudomonas_E fulva | |||||||||||
| CAZyme ID | MGYG000001279_00933 | |||||||||||
| CAZy Family | PL5 | |||||||||||
| CAZyme Description | Alginate lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5225; End: 6328 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL5 | 24 | 340 | 2.5e-174 | 0.9936708860759493 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK00325 | algL | 0.0 | 25 | 363 | 24 | 359 | polysaccharide lyase. |
| cd00244 | AlgLyase | 0.0 | 26 | 359 | 7 | 339 | Alginate Lyase A1-III; enzymatically depolymerizes alginate, a complex copolymer of beta-D-mannuronate and alpha-L-guluronate, by cleaving the beta-(1,4) glycosidic bond. |
| pfam05426 | Alginate_lyase | 3.98e-58 | 60 | 301 | 1 | 274 | Alginate lyase. This family contains several bacterial alginate lyase proteins. Alginate is a family of 1-4-linked copolymers of beta -D-mannuronic acid (M) and alpha -L-guluronic acid (G). It is produced by brown algae and by some bacteria belonging to the genera Azotobacter and Pseudomonas. Alginate lyases catalyze the depolymerization of alginates by beta -elimination, generating a molecule containing 4-deoxy-L-erythro-hex-4-enepyranosyluronate at the nonreducing end. This family adopts an all alpha fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVF54370.1 | 9.32e-276 | 1 | 367 | 1 | 367 |
| QPH49981.1 | 1.32e-275 | 1 | 367 | 1 | 367 |
| QPH44907.1 | 1.32e-275 | 1 | 367 | 1 | 367 |
| CRN05288.1 | 1.32e-275 | 1 | 367 | 1 | 367 |
| QDC05859.1 | 3.80e-275 | 1 | 367 | 1 | 367 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4OZV_A | 3.69e-165 | 24 | 359 | 1 | 334 | CrystalStructure of the periplasmic alginate lyase AlgL [Pseudomonas aeruginosa PAO1] |
| 4OZW_A | 1.22e-163 | 24 | 359 | 1 | 334 | CrystalStructure of the periplasmic alginate lyase AlgL H202A mutant [Pseudomonas aeruginosa PAO1] |
| 7FHZ_A | 1.52e-45 | 48 | 365 | 2 | 306 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 7FHU_A | 1.81e-45 | 48 | 365 | 2 | 306 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHU_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHV_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHV_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHW_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHW_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHX_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHX_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHY_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FHY_B Chain B, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FI0_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a],7FI1_A Chain A, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
| 7FI2_A | 5.31e-44 | 48 | 365 | 2 | 306 | ChainA, Polysaccharide lyase [Stenotrophomonas maltophilia K279a] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B0KGQ9 | 3.51e-240 | 1 | 366 | 1 | 366 | Alginate lyase OS=Pseudomonas putida (strain GB-1) OX=76869 GN=algL PE=3 SV=1 |
| A5W8V4 | 3.91e-237 | 1 | 366 | 1 | 366 | Alginate lyase OS=Pseudomonas putida (strain ATCC 700007 / DSM 6899 / BCRC 17059 / F1) OX=351746 GN=algL PE=3 SV=1 |
| B1J479 | 4.56e-236 | 1 | 366 | 1 | 366 | Alginate lyase OS=Pseudomonas putida (strain W619) OX=390235 GN=algL PE=3 SV=1 |
| Q88ND1 | 5.29e-236 | 1 | 366 | 5 | 370 | Alginate lyase OS=Pseudomonas putida (strain ATCC 47054 / DSM 6125 / CFBP 8728 / NCIMB 11950 / KT2440) OX=160488 GN=algL PE=3 SV=1 |
| Q1I563 | 2.52e-233 | 10 | 366 | 10 | 366 | Alginate lyase OS=Pseudomonas entomophila (strain L48) OX=384676 GN=algL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
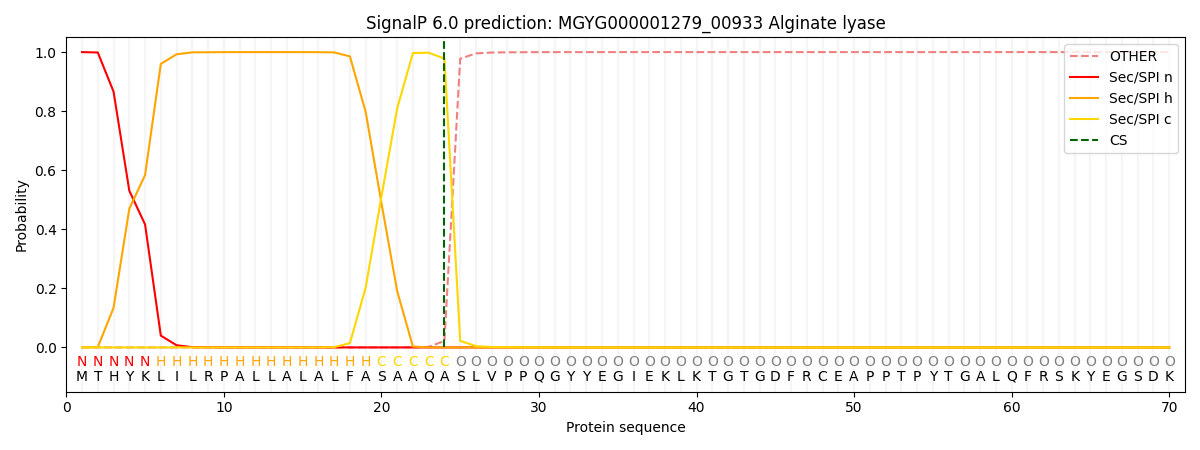
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000228 | 0.999169 | 0.000162 | 0.000154 | 0.000140 | 0.000136 |
