You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001279_01110
You are here: Home > Sequence: MGYG000001279_01110
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudomonas_E fulva | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas_E; Pseudomonas_E fulva | |||||||||||
| CAZyme ID | MGYG000001279_01110 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | Membrane-bound lytic murein transglycosylase F | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1662; End: 3074 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4623 | MltF | 5.21e-113 | 39 | 463 | 14 | 424 | Membrane-bound lytic murein transglycosylase MltF [Cell wall/membrane/envelope biogenesis, Signal transduction mechanisms]. |
| cd01009 | PBP2_YfhD_N | 6.52e-55 | 48 | 289 | 1 | 222 | The solute binding domain of YfhD proteins, a member of the type 2 periplasmic binding fold protein superfamily. This subfamily includes the solute binding domain YfhD_N. These domains are found in the YfhD proteins that are predicted to function as lytic transglycosylases that cleave the glycosidic bond between N-acetylmuramic acid and N-acetylglucosamin in peptidoglycan, while the YfhD_N domain might act as an auxiliary or regulatory subunit. In addition to periplasmic solute binding domain, they have an SLT domain, typically found in soluble lytic transglycosylases, and a C-terminal low complexity domain. The YfhD proteins might have been recruited to create localized cell wall openings required for transport of large substrates such as DNA. They belong to the PBP2 superfamily of periplasmic binding proteins that differ in size and ligand specificity, but have similar tertiary structures consisting of two globular subdomains connected by a flexible hinge. They have been shown to bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. |
| cd13403 | MLTF-like | 5.14e-41 | 311 | 459 | 2 | 161 | membrane-bound lytic murein transglycosylase F (MLTF) and similar proteins. This subfamily includes membrane-bound lytic murein transglycosylase F (MltF, murein lyase F) that degrades murein glycan strands. It is responsible for catalyzing the release of 1,6-anhydromuropeptides from peptidoglycan. Lytic transglycosylase catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do goose-type lysozymes. However, in addition, it also makes a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| PRK10859 | PRK10859 | 5.59e-36 | 1 | 427 | 4 | 407 | membrane-bound lytic murein transglycosylase MltF. |
| cd13707 | PBP2_BvgS_D2 | 1.31e-09 | 48 | 270 | 2 | 207 | The second of the two tandem periplasmic domains of sensor-kinase BvgS; the type 2 peripasmic-binding fold protein. This group contains the second domain of the periplasmic solute-binding domains of BvgS and related proteins. BvgS is composed of two periplasmic domains homologous to bacterial periplasmic-binding proteins (PBPs), a transmembrane region followed successively by a cytoplasmic PAS (Per/ARNT/SIM), a Histidine-kinase (HK), a receiver and a Histidine phosphotransfer (Hpt) domains. The sensor protein BvgS can autophosphorylate and phosphorylate the response regulator BvgA. The BvgAS phosphorelay controls the expression of virulence factors in response to certain environmental stimuli in Bordetella pertussis. Its close homologs, Escherichia coli EvgS and Klebsiella pneumoniae KvgS, appear to be involved in the transcriptional regulation of drug efflux pumps and in countering free radical stresses and sensing iron limiting conditions, respectively. The periplasmic sensor domain of BvgS belongs to the type 2 periplasmic-binding fold protein (PBP2) superfamily, whose members are involved in chemotaxis and uptake of nutrients and other small molecules from the extracellular space as a primary receptor. PBP2 typically comprises of two globular subdomains connected by a flexible hinge and bind their ligand in the cleft between these domains in a manner resembling a Venus flytrap. After binding their specific ligand with high affinity, they can interact with a cognate membrane transport complex comprised of two integral membrane domains and two receptor cytoplasmically-located ATPase domains. This interaction triggers the ligand translocation across the cytoplasmic membrane energized by ATP hydrolysis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDC05095.1 | 0.0 | 1 | 470 | 1 | 470 |
| AVF55114.1 | 0.0 | 1 | 470 | 1 | 470 |
| CRN04527.1 | 0.0 | 1 | 470 | 1 | 470 |
| QPH45674.1 | 0.0 | 1 | 470 | 1 | 470 |
| QPH50760.1 | 0.0 | 1 | 470 | 1 | 470 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AA2_B | 5.12e-24 | 41 | 454 | 45 | 459 | Crystalstructure of MltF from Pseudomonas aeruginosa in complex with NAM-pentapeptide. [Pseudomonas aeruginosa BWHPSA013] |
| 4OYV_A | 1.41e-23 | 41 | 454 | 11 | 425 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with leucine [Pseudomonas aeruginosa PAO1] |
| 5AA3_A | 1.68e-23 | 41 | 454 | 45 | 459 | Crystalstructure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_B Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_C Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_D Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_E Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_F Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_G Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_H Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_I Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_J Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_K Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013],5AA3_L Crystal structure of MltF from Pseudomonas aeruginosa in the presence of tetrasaccharide and tetrapeptide [Pseudomonas aeruginosa BWHPSA013] |
| 4OZ9_A | 2.41e-23 | 41 | 454 | 4 | 418 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with isoleucine [Pseudomonas aeruginosa PAO1] |
| 4OWD_A | 2.57e-23 | 41 | 454 | 11 | 425 | Crystalstructure of MltF from Pseudomonas aeruginosa complexed with cysteine [Pseudomonas aeruginosa PAO1],4OXV_A Crystal structure of MltF from Pseudomonas aeruginosa complexed with valine [Pseudomonas aeruginosa PADK2_CF510],4P0G_A Crystal structure of MltF from Pseudomonas aeruginosa complexed with bulgecin and muropeptide [Pseudomonas aeruginosa PAO1],4P11_A Native crystal structure of MltF Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A4XXV1 | 1.66e-27 | 41 | 454 | 36 | 450 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas mendocina (strain ymp) OX=399739 GN=mltF PE=3 SV=1 |
| B0KRE9 | 2.56e-23 | 41 | 454 | 34 | 448 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas putida (strain GB-1) OX=76869 GN=mltF PE=3 SV=1 |
| Q3KHL5 | 4.68e-23 | 41 | 454 | 34 | 448 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas fluorescens (strain Pf0-1) OX=205922 GN=mltF PE=3 SV=1 |
| A5VZC8 | 6.26e-23 | 41 | 454 | 34 | 448 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas putida (strain ATCC 700007 / DSM 6899 / BCRC 17059 / F1) OX=351746 GN=mltF PE=3 SV=1 |
| Q88P17 | 1.14e-22 | 41 | 454 | 34 | 448 | Membrane-bound lytic murein transglycosylase F OS=Pseudomonas putida (strain ATCC 47054 / DSM 6125 / CFBP 8728 / NCIMB 11950 / KT2440) OX=160488 GN=mltF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
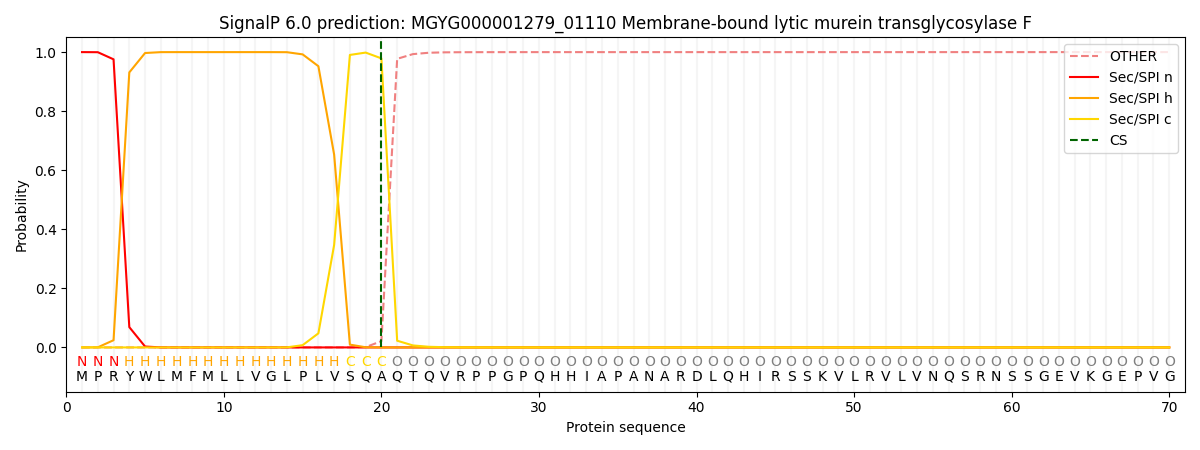
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000188 | 0.999185 | 0.000159 | 0.000152 | 0.000143 | 0.000133 |
