You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001283_00274
You are here: Home > Sequence: MGYG000001283_00274
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Scardovia wiggsiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Scardovia; Scardovia wiggsiae | |||||||||||
| CAZyme ID | MGYG000001283_00274 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 315075; End: 317378 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 60 | 532 | 2.6e-43 | 0.7722222222222223 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13231 | PMT_2 | 6.39e-29 | 114 | 281 | 1 | 159 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1807 | ArnT | 8.36e-28 | 52 | 491 | 1 | 408 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| COG1287 | Stt3 | 7.06e-07 | 113 | 296 | 85 | 264 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
| PRK13279 | arnT | 1.76e-06 | 115 | 282 | 62 | 225 | lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG4745 | COG4745 | 0.003 | 124 | 254 | 74 | 195 | Predicted membrane-bound mannosyltransferase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWZ09319.1 | 1.54e-96 | 64 | 400 | 44 | 380 |
| QUB37647.1 | 7.52e-87 | 52 | 763 | 198 | 792 |
| QHU89858.1 | 1.03e-85 | 50 | 763 | 264 | 901 |
| QHU92512.1 | 4.51e-75 | 50 | 763 | 262 | 898 |
| QHU90800.1 | 6.21e-75 | 50 | 763 | 264 | 898 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34575 | 6.92e-43 | 75 | 749 | 26 | 660 | Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
| P37483 | 7.64e-42 | 64 | 749 | 10 | 647 | Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999881 | 0.000128 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
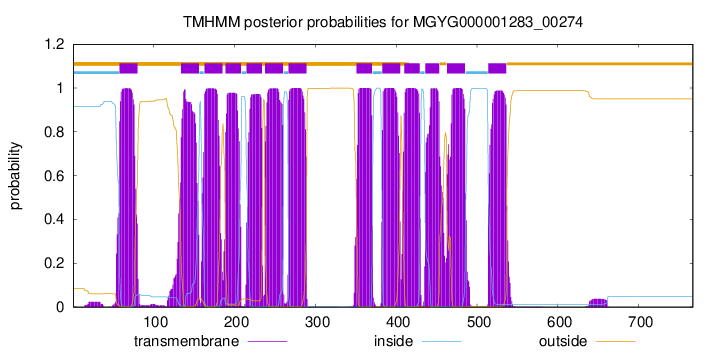
| start | end |
|---|---|
| 58 | 80 |
| 134 | 156 |
| 163 | 185 |
| 189 | 208 |
| 215 | 234 |
| 238 | 260 |
| 267 | 289 |
| 351 | 370 |
| 383 | 405 |
| 410 | 429 |
| 436 | 453 |
| 463 | 485 |
| 514 | 536 |
