You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001283_01188
You are here: Home > Sequence: MGYG000001283_01188
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
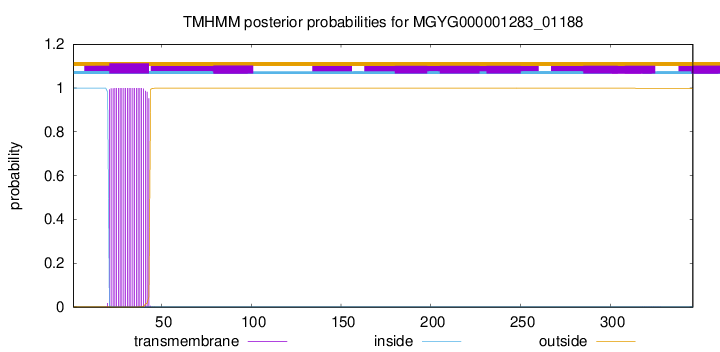
TMHMM annotations
Basic Information help
| Species | Scardovia wiggsiae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Scardovia; Scardovia wiggsiae | |||||||||||
| CAZyme ID | MGYG000001283_01188 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26910; End: 27950 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3583 | YabE | 7.24e-24 | 93 | 228 | 154 | 290 | Uncharacterized conserved protein YabE, contains G5 and tandem DUF348 domains [Function unknown]. |
| pfam07501 | G5 | 1.19e-16 | 149 | 222 | 2 | 75 | G5 domain. This domain is found in a wide range of extracellular proteins. It is found tandemly repeated in up to 8 copies. It is found in the N-terminus of peptidases belonging to the M26 family which cleave human IgA. The domain is also found in proteins involved in metabolism of bacterial cell walls suggesting this domain may have an adhesive function. |
| pfam03990 | DUF348 | 8.13e-06 | 91 | 132 | 1 | 41 | Domain of unknown function (DUF348). This domain normally occurs as tandem repeats; however it is found as a single copy in the S. cerevisiae DNA-binding nuclear protein YCR593. This protein is involved in sporulation part of the SET3C complex, which is required to repress early/middle sporulation genes during meiosis. The bacterial proteins are likely to be involved in a cell wall function as they are found in conjunction with the pfam07501 domain, which is involved in various cell surface processes. |
| cd13399 | Slt35-like | 0.007 | 275 | 305 | 8 | 39 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAR04864.1 | 2.44e-149 | 1 | 346 | 1 | 354 |
| BAR07171.1 | 4.13e-127 | 30 | 346 | 9 | 338 |
| AKV54972.1 | 6.37e-111 | 5 | 346 | 4 | 336 |
| AII74342.1 | 5.09e-109 | 17 | 346 | 16 | 342 |
| AIC91546.1 | 2.05e-108 | 17 | 346 | 16 | 342 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
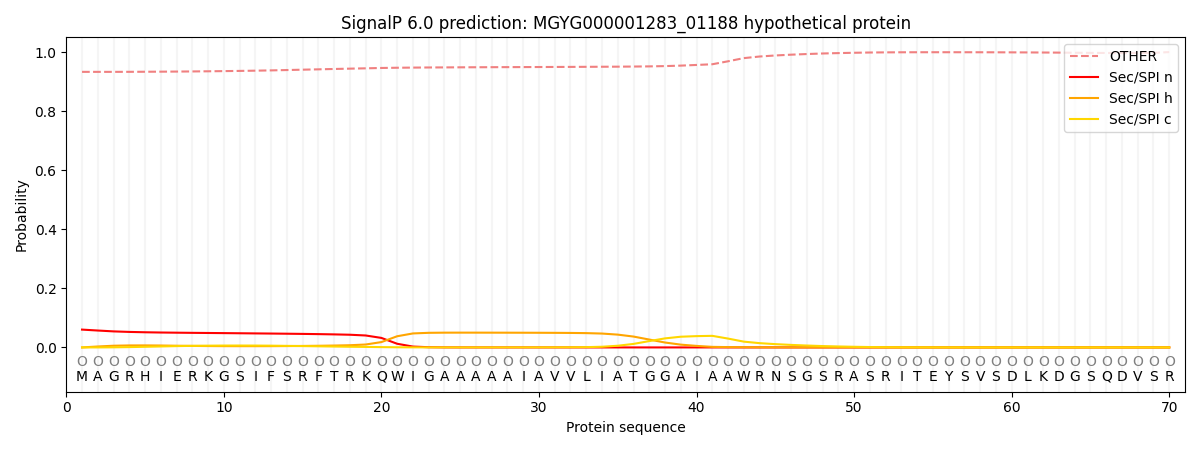
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.935638 | 0.055389 | 0.001174 | 0.000862 | 0.000368 | 0.006581 |