You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001287_00588
You are here: Home > Sequence: MGYG000001287_00588
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bifidobacterium vaginale_G | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium vaginale_G | |||||||||||
| CAZyme ID | MGYG000001287_00588 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35196; End: 38597 Strand: + | |||||||||||
Full Sequence Download help
| MDSITAHNLT FEDEVLQVLS SAKDAHPHDV DSSIMAMMCV ERDTRFYADT LHAVLSQSIL | 60 |
| PGTLVIVDCA SRVKQTMQTN LQIKINSSKL AGAINTRKSL LQNANNIDSN NAIDSDSKYR | 120 |
| PNTNRVNNTR VNNSENKSVH IIIIPISYAH SFGDAIEKSL RKILPCAGVS ALWLLHDDSR | 180 |
| PADFHCLEYL RETWRNTPTA SVLGAKQVDW QGNILHNVGM YAWRHGVHTL TVDGEPDQEQ | 240 |
| YDGRGDVYAV SLAGALVTLT AWQTLRGTQQ WMTTFGESRD FCRRVCLSGG RVVVVPSARI | 300 |
| AHRRARLEGI RTRNGDARNH NNSGSKYGVA ASLVAKQRYK YTDISIFLWP LIWILSLPAC | 360 |
| FVGALHSLFA KHPYIAWIRL LLPWLAIMQL PRAVFARRRV ARNTRVPLKR LVPLIADRHQ | 420 |
| VARWRDRSSA FQAQQHFVLL SPLAKRHLHI RVLRRWSAVI LASLLLFAVI LNLYGSPWRE | 480 |
| ILSGASWNAQ QWLPTGADFN QLWNSAIGAW VPDDGFGPAI PPAPWLSIWA IASIVVGANP | 540 |
| LLAVTLMFFL AAPAMCLSFW ALAGVFTRSD TVRVCAGICW TMIAIAFGLF ARGDLPMLMT | 600 |
| MAFLPAAFAF AFHAVGMYVT EDPVRPVKST QCAACAALCF MVPVAAEPQL VLPLIVIFIA | 660 |
| SFIMVRSHRI MFALMPIPSI IVLLPTIGSA IRYGESGMWR QLFSSMALPL RSVQGAPRVS | 720 |
| DYADVLLHAF NMASPNVEIN MPLMSLRTII MVIFAILAVV MAVTSLTLPF ALRVSRMMWV | 780 |
| VIISGMLLSL IAARVVVASD ISGPVAASVL PGVALSTVGI LSCMCMVAGQ AVRRFEPLRT | 840 |
| SKESESQIFD NYKTRASRRA AMRRSAAQLL QRIIRIARAV LASSMLTMAV LLGLFAVLSG | 900 |
| PATSICANNN ELPMVVSDYL RKDDTRRILA LRAQSSHDLS ISIMRTSKGD AIDESPALRV | 960 |
| RQVMFGDSSS NKAIYRASAQ LLSHADNGAI STLKQLGIGG IYIVNSSENK QVSHKTSMSI | 1020 |
| QQLLHPSVQT VSNVNKEATE QLIAHVNASD GTQLVVSSPQ GTYCRFDGVP DSKSLSSTSP | 1080 |
| YYRSRPLAWR VPWIIVSSMV LLLYCIIAVP RFGRHAMIER SDDRHEEELD ELA | 1133 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13632 | Glyco_trans_2_3 | 3.84e-06 | 174 | 390 | 4 | 193 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APW18624.1 | 0.0 | 1 | 1133 | 1 | 1133 |
| AYZ22105.1 | 0.0 | 1 | 1123 | 1 | 1099 |
| BAQ33468.1 | 0.0 | 1 | 1123 | 1 | 1099 |
| SDR71691.1 | 0.0 | 1 | 1123 | 1 | 1099 |
| VEH17710.1 | 0.0 | 1 | 1123 | 1 | 1099 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000056 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
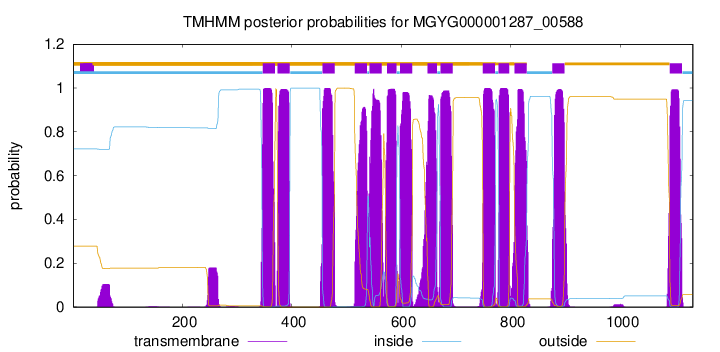
| start | end |
|---|---|
| 347 | 369 |
| 374 | 396 |
| 456 | 478 |
| 515 | 537 |
| 542 | 564 |
| 574 | 591 |
| 598 | 620 |
| 648 | 665 |
| 672 | 694 |
| 749 | 771 |
| 778 | 797 |
| 807 | 829 |
| 876 | 898 |
| 1091 | 1113 |
