You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001287_00727
You are here: Home > Sequence: MGYG000001287_00727
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
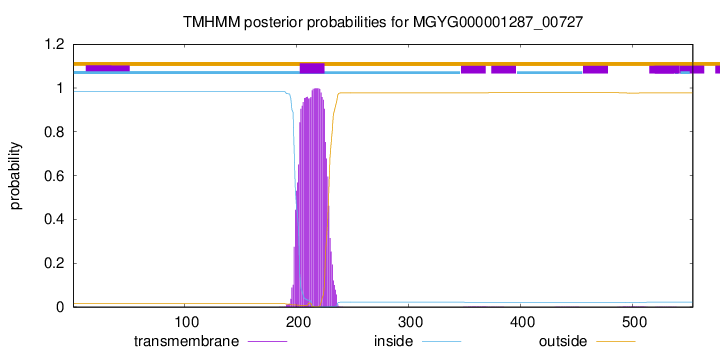
TMHMM annotations
Basic Information help
| Species | Bifidobacterium vaginale_G | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Bifidobacteriaceae; Bifidobacterium; Bifidobacterium vaginale_G | |||||||||||
| CAZyme ID | MGYG000001287_00727 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49197; End: 50861 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 264 | 396 | 2e-24 | 0.8592592592592593 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd13399 | Slt35-like | 3.59e-33 | 265 | 389 | 3 | 107 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| PRK08581 | PRK08581 | 5.46e-27 | 440 | 526 | 497 | 592 | amidase domain-containing protein. |
| COG3942 | COG3942 | 8.41e-27 | 370 | 554 | 2 | 171 | Surface antigen [Cell wall/membrane/envelope biogenesis]. |
| cd00254 | LT-like | 7.81e-24 | 267 | 389 | 1 | 111 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| pfam05257 | CHAP | 1.26e-19 | 441 | 526 | 4 | 82 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTL78627.1 | 3.59e-136 | 146 | 554 | 176 | 574 |
| ALE11699.1 | 2.28e-134 | 146 | 554 | 176 | 574 |
| AFL04801.1 | 9.07e-134 | 146 | 554 | 176 | 574 |
| CDL13121.2 | 9.07e-134 | 146 | 554 | 176 | 574 |
| QRI58211.1 | 9.07e-134 | 146 | 554 | 176 | 574 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T1Q_A | 1.03e-15 | 446 | 553 | 252 | 356 | ChainA, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_B Chain B, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_C Chain C, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325],5T1Q_D Chain D, N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 2K3A_A | 4.37e-10 | 442 | 554 | 52 | 153 | ChainA, CHAP domain protein [Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 = NCTC 7292] |
| 2LRJ_A | 1.47e-09 | 442 | 554 | 11 | 112 | ChainA, Staphyloxanthin biosynthesis protein, putative [Staphylococcus aureus subsp. aureus COL] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31976 | 1.63e-14 | 265 | 385 | 1435 | 1538 | SPbeta prophage-derived uncharacterized transglycosylase YomI OS=Bacillus subtilis (strain 168) OX=224308 GN=yomI PE=3 SV=2 |
| O64046 | 1.63e-14 | 265 | 385 | 1435 | 1538 | Probable tape measure protein OS=Bacillus phage SPbeta OX=66797 GN=yomI PE=3 SV=1 |
| Q2G222 | 1.64e-14 | 446 | 553 | 512 | 616 | N-acetylmuramoyl-L-alanine amidase domain-containing protein SAOUHSC_02979 OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=SAOUHSC_02979 PE=1 SV=1 |
| O31608 | 1.91e-12 | 265 | 384 | 73 | 175 | Putative murein lytic transglycosylase YjbJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yjbJ PE=3 SV=1 |
| Q8CMN2 | 1.10e-11 | 446 | 554 | 224 | 324 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=sle1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
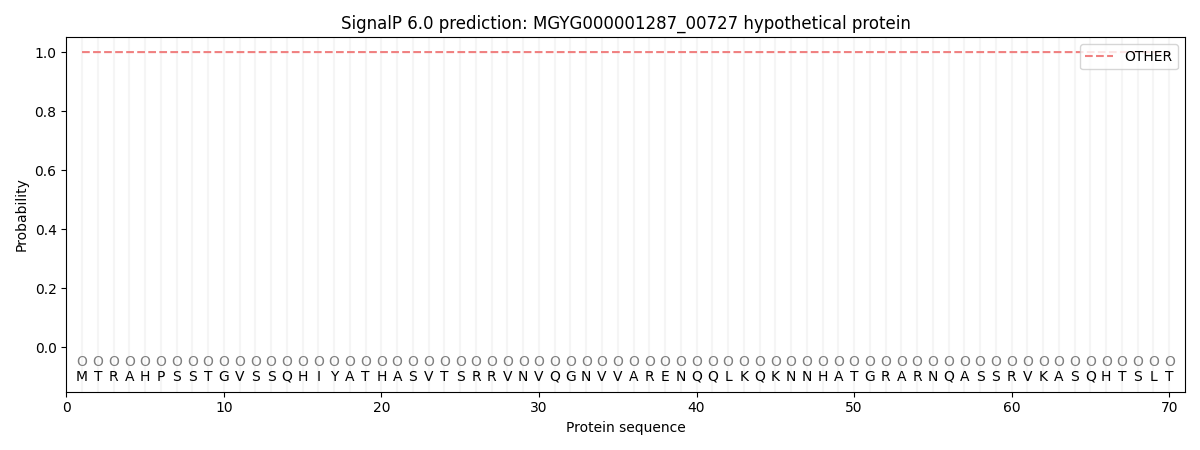
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000041 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |