You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001289_01451
You are here: Home > Sequence: MGYG000001289_01451
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Streptococcus sp900543065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Streptococcus; Streptococcus sp900543065 | |||||||||||
| CAZyme ID | MGYG000001289_01451 | |||||||||||
| CAZy Family | GH25 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3353; End: 7135 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH25 | 138 | 324 | 7.8e-41 | 0.9717514124293786 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06522 | GH25_AtlA-like | 1.13e-75 | 135 | 338 | 1 | 192 | AtlA is an autolysin found in Gram-positive lactic acid bacteria that degrades bacterial cell walls by catalyzing the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues. This family includes the AtlA and Aml autolysins from Streptococcus mutans which have a C-terminal glycosyl hydrolase family 25 (GH25) catalytic domain as well as six tandem N-terminal repeats of the GBS (group B Streptococcus) Bsp-like peptidoglycan-binding domain. Other members of this family have one or more C-terminal peptidoglycan-binding domain(s) (SH3 or LysM) in addition to the GH25 domain. |
| pfam01183 | Glyco_hydro_25 | 1.33e-37 | 138 | 322 | 1 | 174 | Glycosyl hydrolases family 25. |
| pfam08481 | GBS_Bsp-like | 7.85e-31 | 884 | 972 | 1 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 1.71e-30 | 363 | 451 | 1 | 89 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
| pfam08481 | GBS_Bsp-like | 1.03e-26 | 575 | 662 | 1 | 88 | GBS Bsp-like repeat. This domain is found as a repeat in a number of Streptococcus proteins including some hypothetical proteins and Bsp. Bsp is a protein of group B Streptococcus (GBS) which might control cell morphology. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKH69741.1 | 0.0 | 127 | 1260 | 153 | 1286 |
| VED88051.1 | 0.0 | 1 | 1260 | 1 | 1280 |
| QUB38816.1 | 0.0 | 1 | 1260 | 1 | 1155 |
| QEW10542.1 | 0.0 | 1 | 1260 | 1 | 1156 |
| ARC22445.1 | 0.0 | 127 | 1260 | 148 | 1183 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
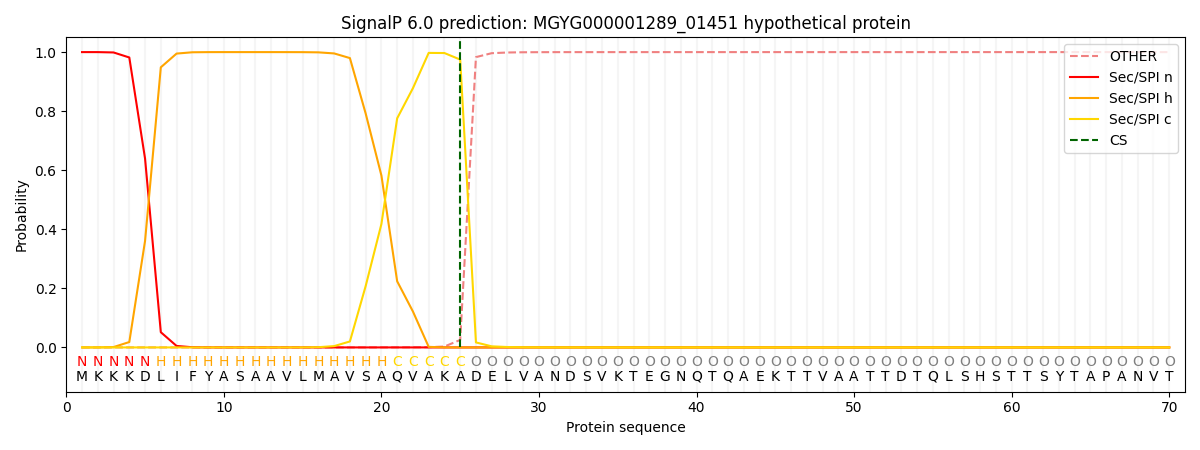
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000317 | 0.998859 | 0.000300 | 0.000173 | 0.000175 | 0.000152 |
