You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001296_01571
You are here: Home > Sequence: MGYG000001296_01571
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pediococcus acidilactici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Pediococcus; Pediococcus acidilactici | |||||||||||
| CAZyme ID | MGYG000001296_01571 | |||||||||||
| CAZy Family | AA10 | |||||||||||
| CAZyme Description | GlcNAc-binding protein A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1614368; End: 1614967 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA10 | 33 | 197 | 3.6e-49 | 0.9943820224719101 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13211 | PRK13211 | 3.70e-70 | 30 | 198 | 21 | 193 | N-acetylglucosamine-binding protein GbpA. |
| cd21177 | LPMO_AA10 | 1.65e-69 | 33 | 197 | 1 | 180 | lytic polysaccharide monooxygenase (LPMO) auxiliary activity family 10 (AA10). AA10 proteins are copper-dependent lytic polysaccharide monooxygenases (LPMOs), which may act on chitin or cellulose. The family used to be called CBM33. Activities in this family include lytic cellulose monooxygenase (C1-hydroxylating) (EC 1.14.99.54), lytic cellulose monooxygenase (C4-dehydrogenating) (EC 1.14.99.56), lytic chitin monooxygenase (EC 1.14.99.53), and lytic xylan monooxygenase/xylan oxidase (glycosidic bond-cleaving) (EC 1.14.99.-). Also included are viral chitin-binding glycoproteins such as fusolin and spheroidin-like proteins. |
| COG3397 | COG3397 | 1.01e-65 | 27 | 198 | 24 | 207 | Predicted carbohydrate-binding protein, contains CBM5 and CBM33 domains [General function prediction only]. |
| pfam03067 | LPMO_10 | 2.04e-57 | 33 | 196 | 1 | 186 | Lytic polysaccharide mono-oxygenase, cellulose-degrading. This domain is found associated with a wide variety of cellulose binding domains. This is a family of two very closely related proteins that together act as both a C1- and a C4-oxidising lytic polysaccharide mono-oxygenase, degrading cellulose. This domain is also found in baculoviral spheroidins and spindolins, protein of unknown function. |
| cd21174 | LPMO_auxiliary | 7.68e-08 | 95 | 195 | 35 | 136 | lytic polysaccharide monooxygenase auxiliary activity protein. Many proteins in this superfamily are copper-dependent lytic polysaccharide monooxygenases (LPMOs) and include lytic polysaccharide monooxygenase auxiliary activity families 9 (AA9) and 10 (AA10). The substrate-binding surface of this family is a flat beta-sandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHM51471.1 | 9.99e-146 | 1 | 199 | 1 | 199 |
| QHS02361.1 | 9.99e-146 | 1 | 199 | 1 | 199 |
| AZP90285.1 | 9.99e-146 | 1 | 199 | 1 | 199 |
| QHM54277.1 | 9.99e-146 | 1 | 199 | 1 | 199 |
| QYI95284.1 | 5.78e-145 | 1 | 199 | 1 | 199 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4A02_A | 9.77e-71 | 33 | 199 | 1 | 166 | X-raycrystallographic structure of EfCBM33A [Enterococcus faecalis],4ALC_A X-Ray photoreduction of Polysaccharide monooxigenase CBM33 [Enterococcus faecalis],4ALE_A Structure changes of Polysaccharide monooxygenase CBM33A from Enterococcus faecalis by X-ray induced photoreduction. [Enterococcus faecalis],4ALQ_A X-Ray photoreduction of Polysaccharide monooxygenase CBM33 [Enterococcus faecalis V583],4ALR_A X-Ray photoreduction of Polysaccharide monooxygenase CBM33 [Enterococcus faecalis],4ALS_A X-Ray photoreduction of Polysaccharide monooxygenase CBM33 [Enterococcus faecalis],4ALT_A X-Ray photoreduction of Polysaccharide monooxygenase CBM33 [Enterococcus faecalis] |
| 5L2V_A | 2.10e-55 | 33 | 198 | 1 | 164 | Catalyticdomain of LPMO Lmo2467 from Listeria monocytogenes [Listeria monocytogenes 10403S],5L2V_B Catalytic domain of LPMO Lmo2467 from Listeria monocytogenes [Listeria monocytogenes 10403S] |
| 5WSZ_A | 3.50e-52 | 33 | 198 | 1 | 166 | Crystalstructure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_B Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_C Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki],5WSZ_D Crystal structure of a lytic polysaccharide monooxygenase,BtLPMO10A, from Bacillus thuringiensis [Bacillus thuringiensis serovar kurstaki] |
| 2BEM_A | 2.37e-50 | 33 | 198 | 1 | 168 | Crystalstructure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2BEM_B Crystal structure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2BEM_C Crystal structure of the Serratia marcescens chitin-binding protein CBP21 [Serratia marcescens],2LHS_A Structure of the chitin binding protein 21 (CBP21) [Serratia marcescens] |
| 5LW4_A | 7.17e-50 | 33 | 198 | 1 | 169 | NMRsolution structure of the apo-form of the chitin-active lytic polysaccharide monooxygenase BlLPMO10A [Bacillus licheniformis],6TWE_A Cu(I) NMR solution structure of the chitin-active lytic polysaccharide monooxygenase BlLPMO10A [Bacillus licheniformis DSM 13 = ATCC 14580] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q838S1 | 2.33e-70 | 30 | 199 | 26 | 194 | Lytic chitin monooxygenase OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0362 PE=1 SV=1 |
| Q87FT0 | 1.03e-37 | 32 | 196 | 23 | 199 | GlcNAc-binding protein A OS=Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633) OX=223926 GN=gbpA PE=3 SV=1 |
| Q6LKG5 | 2.80e-37 | 21 | 196 | 13 | 200 | GlcNAc-binding protein A OS=Photobacterium profundum (strain SS9) OX=298386 GN=gbpA PE=3 SV=1 |
| Q8EHY2 | 3.29e-37 | 30 | 197 | 25 | 194 | GlcNAc-binding protein A OS=Shewanella oneidensis (strain MR-1) OX=211586 GN=gbpA PE=3 SV=2 |
| Q8D7V4 | 5.24e-36 | 56 | 196 | 52 | 198 | GlcNAc-binding protein A OS=Vibrio vulnificus (strain CMCP6) OX=216895 GN=gbpA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
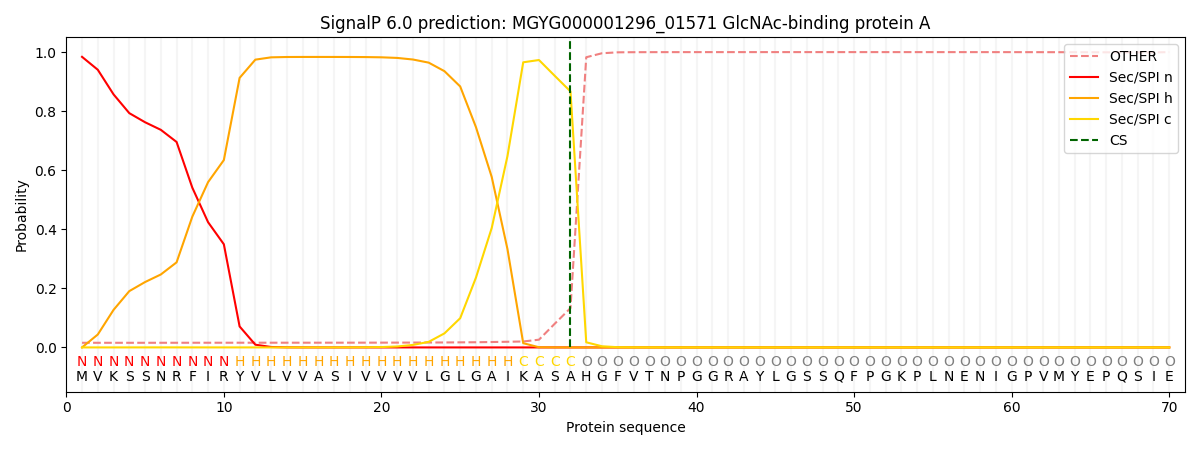
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.022708 | 0.974979 | 0.001239 | 0.000398 | 0.000323 | 0.000326 |

