You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001297_00792
You are here: Home > Sequence: MGYG000001297_00792
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Campylobacter_D coli | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Campylobacteraceae; Campylobacter_D; Campylobacter_D coli | |||||||||||
| CAZyme ID | MGYG000001297_00792 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 781806; End: 782537 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2992 | Bax | 4.90e-85 | 1 | 239 | 26 | 261 | Uncharacterized FlgJ-related protein [General function prediction only]. |
| PRK10356 | PRK10356 | 4.08e-18 | 107 | 230 | 145 | 264 | protein bax. |
| pfam01832 | Glucosaminidase | 9.97e-11 | 111 | 175 | 13 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| COG1705 | FlgJ | 1.49e-07 | 110 | 242 | 62 | 192 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| smart00047 | LYZ2 | 9.20e-07 | 107 | 193 | 24 | 116 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOP67625.1 | 5.45e-174 | 1 | 243 | 1 | 243 |
| QGG38450.1 | 5.45e-174 | 1 | 243 | 1 | 243 |
| QUJ27183.1 | 5.45e-174 | 1 | 243 | 1 | 243 |
| QLD91513.1 | 5.45e-174 | 1 | 243 | 1 | 243 |
| QYH13551.1 | 5.45e-174 | 1 | 243 | 1 | 243 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27297 | 3.88e-13 | 106 | 230 | 144 | 264 | Protein bax OS=Escherichia coli (strain K12) OX=83333 GN=bax PE=4 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
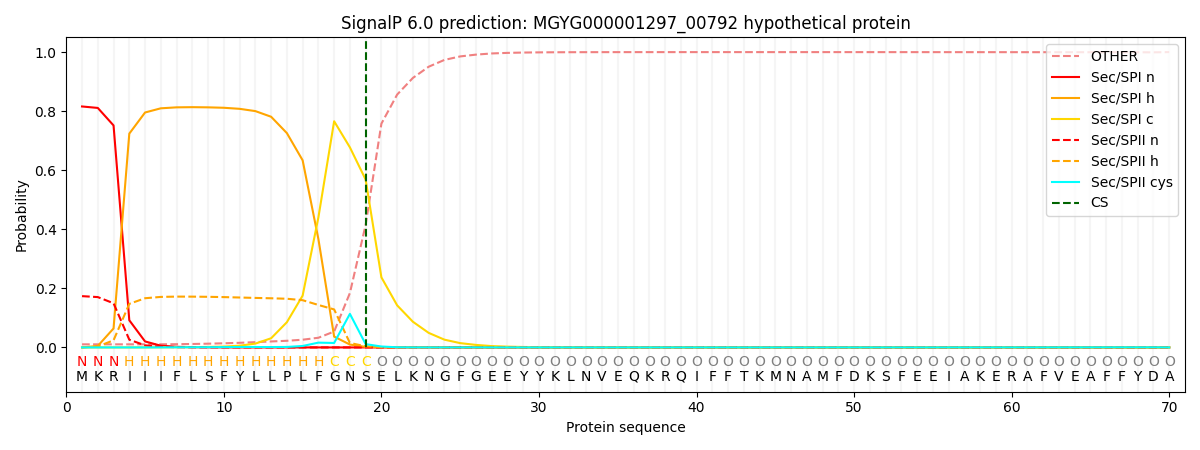
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.012268 | 0.809898 | 0.177038 | 0.000273 | 0.000260 | 0.000258 |
