You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001298_01922
You are here: Home > Sequence: MGYG000001298_01922
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
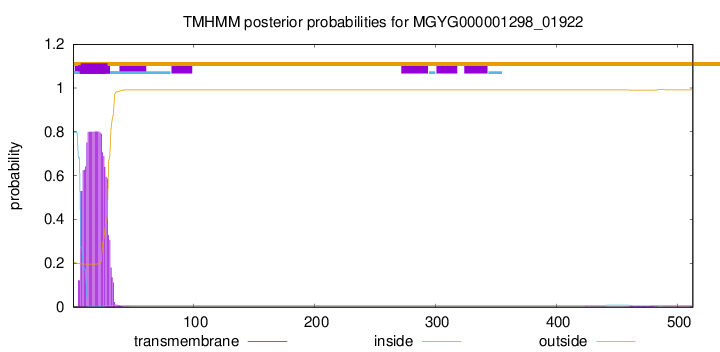
TMHMM annotations
Basic Information help
| Species | Listeria grayi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria grayi | |||||||||||
| CAZyme ID | MGYG000001298_01922 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Cell division suppressor protein YneA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 408397; End: 409938 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK13914 | PRK13914 | 3.50e-154 | 1 | 510 | 3 | 481 | invasion associated endopeptidase. |
| pfam00877 | NLPC_P60 | 9.15e-45 | 407 | 509 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| COG0791 | Spr | 3.46e-25 | 398 | 507 | 78 | 196 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| PRK06347 | PRK06347 | 1.34e-22 | 22 | 341 | 326 | 592 | 1,4-beta-N-acetylmuramoylhydrolase. |
| NF033742 | NlpC_p60_RipB | 1.80e-18 | 406 | 492 | 88 | 186 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AEO72020.1 | 4.87e-292 | 1 | 510 | 3 | 512 |
| VEI32232.1 | 9.71e-290 | 1 | 513 | 1 | 513 |
| QDA73944.1 | 2.52e-137 | 1 | 510 | 1 | 522 |
| AEO72015.1 | 2.68e-137 | 1 | 510 | 3 | 524 |
| SNV19148.1 | 1.43e-136 | 1 | 510 | 1 | 522 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6B8C_A | 2.16e-20 | 397 | 492 | 30 | 126 | Crystalstructure of NlpC/p60 domain of peptidoglycan hydrolase SagA [Enterococcus faecium] |
| 7CFL_A | 9.09e-19 | 402 | 510 | 21 | 137 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 4HPE_A | 8.06e-14 | 401 | 510 | 193 | 306 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 3NE0_A | 2.69e-12 | 406 | 492 | 96 | 194 | Structureand functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation [Mycobacterium tuberculosis H37Rv] |
| 3PBC_A | 2.69e-12 | 406 | 492 | 96 | 194 | ChainA, Invasion Protein [Mycobacterium tuberculosis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q01835 | 1.81e-283 | 1 | 510 | 3 | 511 | Probable endopeptidase p60 OS=Listeria grayi OX=1641 GN=iap PE=3 SV=1 |
| Q01839 | 3.04e-137 | 1 | 510 | 3 | 524 | Probable endopeptidase p60 OS=Listeria welshimeri OX=1643 GN=iap PE=3 SV=1 |
| Q01837 | 7.80e-135 | 1 | 510 | 3 | 524 | Probable endopeptidase p60 OS=Listeria ivanovii OX=1638 GN=iap PE=3 SV=1 |
| Q01838 | 2.14e-134 | 1 | 510 | 3 | 523 | Probable endopeptidase p60 OS=Listeria seeligeri OX=1640 GN=iap PE=3 SV=1 |
| P21171 | 9.77e-114 | 1 | 510 | 3 | 484 | Probable endopeptidase p60 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=iap PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
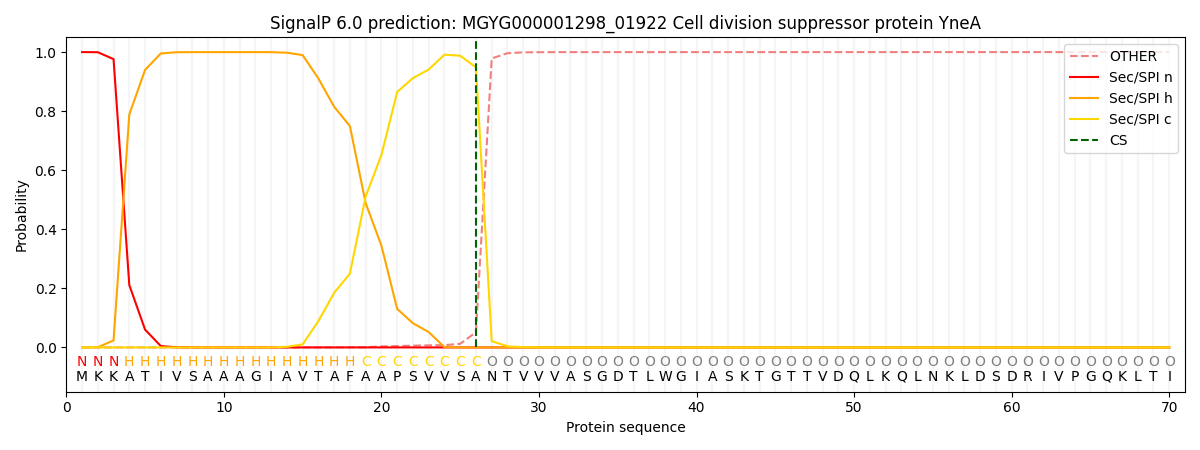
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000268 | 0.998989 | 0.000169 | 0.000191 | 0.000184 | 0.000154 |