You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001298_02463
You are here: Home > Sequence: MGYG000001298_02463
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
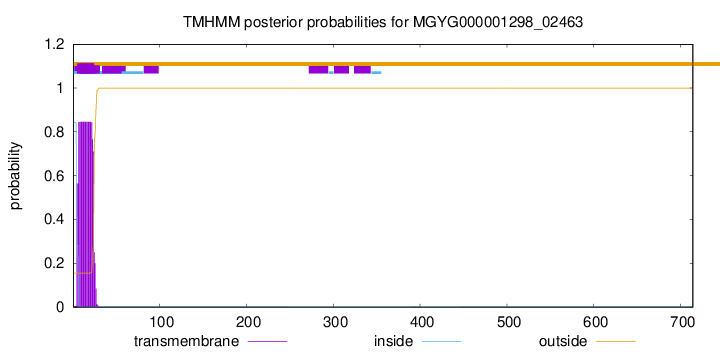
TMHMM annotations
Basic Information help
| Species | Listeria grayi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria grayi | |||||||||||
| CAZyme ID | MGYG000001298_02463 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 953104; End: 955248 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 575 | 710 | 3.2e-16 | 0.675531914893617 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd07473 | Peptidases_S8_Subtilisin_like | 4.65e-87 | 154 | 399 | 1 | 258 | Peptidase S8 family domain in Subtilisin-like proteins. This family is a member of the Peptidases S8 or Subtilases serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| cd07477 | Peptidases_S8_Subtilisin_subset | 1.20e-61 | 157 | 398 | 2 | 229 | Peptidase S8 family domain in Subtilisin proteins. This group is composed of many different subtilisins: Pro-TK-subtilisin, subtilisin Carlsberg, serine protease Pb92 subtilisin, and BPN subtilisins just to name a few. Pro-TK-subtilisin is a serine protease from the hyperthermophilic archaeon Thermococcus kodakaraensis and consists of a signal peptide, a propeptide, and a mature domain. TK-subtilisin is matured from pro-TK-subtilisin upon autoprocessing and degradation of the propeptide. Unlike other subtilisins though, the folding of the unprocessed form of pro-TK-subtilisin is induced by Ca2+ binding which is almost completed prior to autoprocessing. Ca2+ is required for activity unlike the bacterial subtilisins. The propeptide is not required for folding of the mature domain unlike the bacterial subtilases because of the stability produced from Ca2+ binding. Subtilisin Carlsberg is extremely similar in structure to subtilisin BPN'/Novo thought it has a 30% difference in amino acid sequence. The substrate binding regions are also similar and 2 possible Ca2+ binding sites have been identified recently. Subtilisin Carlsberg possesses the highest commercial importance as a proteolytic additive for detergents. Serine protease Pb92, the serine protease from the alkalophilic Bacillus strain PB92, also contains two calcium ions and the overall folding of the polypeptide chain closely resembles that of the subtilisins. Members of the peptidases S8 and S35 clan include endopeptidases, exopeptidases and also a tripeptidyl-peptidase. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The S53 family contains a catalytic triad Glu/Asp/Ser. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
| cd07484 | Peptidases_S8_Thermitase_like | 9.11e-59 | 134 | 403 | 2 | 260 | Peptidase S8 family domain in Thermitase-like proteins. Thermitase is a non-specific, trypsin-related serine protease with a very high specific activity. It contains a subtilisin like domain. The tertiary structure of thermitase is similar to that of subtilisin BPN'. It contains a Asp/His/Ser catalytic triad. Members of the peptidases S8 (subtilisin and kexin) and S53 (sedolisin) clan include endopeptidases and exopeptidases. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. Serine acts as a nucleophile, aspartate as an electrophile, and histidine as a base. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of subtilisin. The serine residue here is the nucleophilic equivalent of the serine residue in the S8 family, while glutamic acid has the same role here as the histidine base. However, the aspartic acid residue that acts as an electrophile is quite different. In S53 the it follows glutamic acid, while in S8 it precedes histidine. The stability of these enzymes may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. There is a great diversity in the characteristics of their members: some contain disulfide bonds, some are intracellular while others are extracellular, some function at extreme temperatures, and others at high or low pH values. |
| cd00306 | Peptidases_S8_S53 | 9.99e-54 | 157 | 398 | 1 | 241 | Peptidase domain in the S8 and S53 families. Members of the peptidases S8 (subtilisin and kexin) and S53 (sedolisin) family include endopeptidases and exopeptidases. The S8 family has an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. Serine acts as a nucleophile, aspartate as an electrophile, and histidine as a base. The S53 family contains a catalytic triad Glu/Asp/Ser with an additional acidic residue Asp in the oxyanion hole, similar to that of subtilisin. The serine residue here is the nucleophilic equivalent of the serine residue in the S8 family, while glutamic acid has the same role here as the histidine base. However, the aspartic acid residue that acts as an electrophile is quite different. In S53, it follows glutamic acid, while in S8 it precedes histidine. The stability of these enzymes may be enhanced by calcium; some members have been shown to bind up to 4 ions via binding sites with different affinity. There is a great diversity in the characteristics of their members: some contain disulfide bonds, some are intracellular while others are extracellular, some function at extreme temperatures, and others at high or low pH values. |
| cd07474 | Peptidases_S8_subtilisin_Vpr-like | 4.97e-45 | 155 | 420 | 2 | 295 | Peptidase S8 family domain in Vpr-like proteins. The maturation of the peptide antibiotic (lantibiotic) subtilin in Bacillus subtilis ATCC 6633 includes posttranslational modifications of the propeptide and proteolytic cleavage of the leader peptide. Vpr was identified as one of the proteases, along with WprA, that are capable of processing subtilin. Asp, Ser, His triadPeptidases S8 or Subtilases are a serine endo- and exo-peptidase clan. They have an Asp/His/Ser catalytic triad similar to that found in trypsin-like proteases, but do not share their three-dimensional structure and are not homologous to trypsin. The stability of subtilases may be enhanced by calcium, some members have been shown to bind up to 4 ions via binding sites with different affinity. Some members of this clan contain disulfide bonds. These enzymes can be intra- and extracellular, some function at extreme temperatures and pH values. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEI36172.1 | 0.0 | 1 | 714 | 1 | 714 |
| BAY62672.1 | 7.05e-49 | 154 | 421 | 66 | 349 |
| QIR40666.1 | 7.73e-49 | 152 | 428 | 61 | 358 |
| BAY27877.1 | 9.78e-49 | 149 | 421 | 57 | 350 |
| BAY12514.1 | 1.57e-48 | 149 | 421 | 172 | 443 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1DBI_A | 5.06e-37 | 158 | 420 | 34 | 277 | CrystalStructure Of A Thermostable Serine Protease [Bacillus sp. Ak1] |
| 3VV3_A | 2.16e-35 | 155 | 420 | 42 | 324 | Crystalstructure of deseasin MCP-01 from Pseudoalteromonas sp. SM9913 [Pseudoalteromonas sp. SM9913],3VV3_B Crystal structure of deseasin MCP-01 from Pseudoalteromonas sp. SM9913 [Pseudoalteromonas sp. SM9913] |
| 1AK9_A | 1.66e-34 | 157 | 402 | 26 | 257 | ChainA, SUBTILISIN 8321 [Bacillus amyloliquefaciens] |
| 1S01_A | 2.26e-34 | 157 | 402 | 26 | 257 | LARGEINCREASES IN GENERAL STABILITY FOR SUBTILISIN BPN(PRIME) THROUGH INCREMENTAL CHANGES IN THE FREE ENERGY OF UNFOLDING [Bacillus amyloliquefaciens] |
| 1AQN_A | 2.26e-34 | 157 | 402 | 26 | 257 | ChainA, SUBTILISIN 8324 [Bacillus amyloliquefaciens],1AU9_A Chain A, SUBTILISIN BPN' [Bacillus amyloliquefaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q45670 | 2.08e-37 | 10 | 420 | 8 | 398 | Thermophilic serine proteinase OS=Bacillus sp. (strain AK1) OX=268807 PE=1 SV=1 |
| P00781 | 1.71e-31 | 157 | 395 | 26 | 249 | Subtilisin DY OS=Bacillus licheniformis OX=1402 GN=apr PE=1 SV=1 |
| P00782 | 6.45e-31 | 157 | 402 | 133 | 364 | Subtilisin BPN' OS=Bacillus amyloliquefaciens OX=1390 GN=apr PE=1 SV=1 |
| P11018 | 2.18e-30 | 155 | 380 | 42 | 260 | Major intracellular serine protease OS=Bacillus subtilis (strain 168) OX=224308 GN=isp PE=1 SV=2 |
| P00780 | 1.26e-29 | 157 | 395 | 131 | 354 | Subtilisin Carlsberg OS=Bacillus licheniformis OX=1402 GN=subC PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
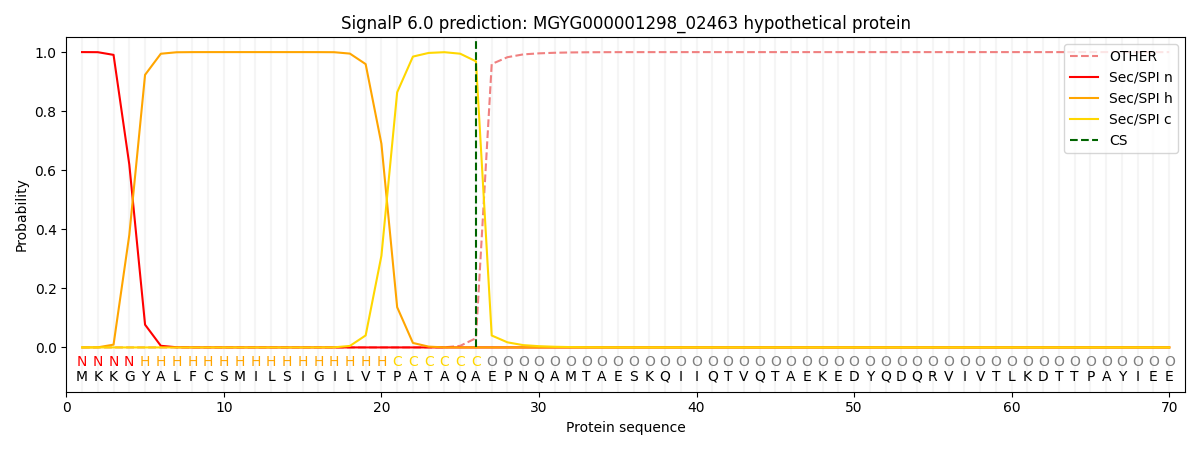
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000269 | 0.999033 | 0.000179 | 0.000188 | 0.000167 | 0.000150 |