You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001298_02589
You are here: Home > Sequence: MGYG000001298_02589
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
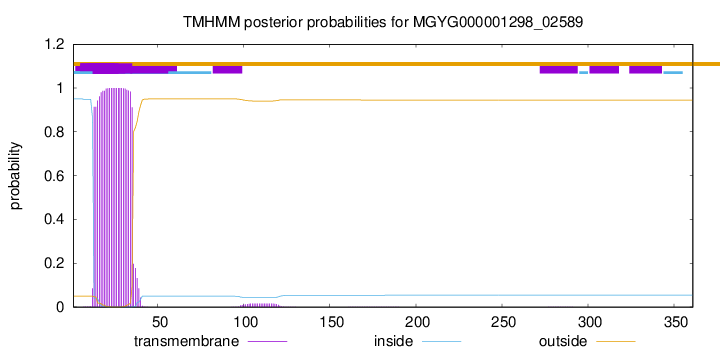
TMHMM annotations
Basic Information help
| Species | Listeria grayi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Listeriaceae; Listeria; Listeria grayi | |||||||||||
| CAZyme ID | MGYG000001298_02589 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37301; End: 38386 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 75 | 213 | 3.4e-20 | 0.8740740740740741 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00877 | NLPC_P60 | 2.65e-32 | 245 | 356 | 3 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| cd13399 | Slt35-like | 2.51e-24 | 77 | 208 | 1 | 108 | Slt35-like lytic transglycosylase. Lytic transglycosylase similar to Escherichia coli lytic transglycosylase Slt35 and Pseudomonas aeruginosa Sltb1. Lytic transglycosylase (LT) catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| COG0791 | Spr | 2.52e-21 | 166 | 353 | 15 | 195 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
| PRK10838 | spr | 1.70e-15 | 250 | 359 | 85 | 185 | bifunctional murein DD-endopeptidase/murein LD-carboxypeptidase. |
| NF033742 | NlpC_p60_RipB | 3.04e-14 | 215 | 336 | 62 | 184 | NlpC/P60 family peptidoglycan endopeptidase RipB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGT06905.1 | 7.01e-268 | 1 | 361 | 1 | 361 |
| AWW22405.1 | 7.90e-247 | 29 | 361 | 2 | 334 |
| QAT55802.1 | 3.34e-150 | 2 | 359 | 13 | 380 |
| ASK26302.1 | 9.52e-150 | 2 | 359 | 13 | 380 |
| QSF36411.1 | 1.67e-142 | 34 | 358 | 27 | 357 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2K1G_A | 7.76e-10 | 250 | 359 | 24 | 124 | SolutionNMR structure of lipoprotein spr from Escherichia coli K12. Northeast Structural Genomics target ER541-37-162 [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40767 | 1.10e-55 | 230 | 359 | 343 | 472 | Peptidoglycan DL-endopeptidase CwlO OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlO PE=1 SV=2 |
| Q65NQ9 | 1.92e-55 | 218 | 359 | 312 | 451 | Peptidoglycan DL-endopeptidase CwlO OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=cwlO PE=3 SV=1 |
| O07532 | 8.59e-12 | 245 | 356 | 387 | 486 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| P54421 | 1.61e-11 | 244 | 356 | 233 | 333 | Probable peptidoglycan endopeptidase LytE OS=Bacillus subtilis (strain 168) OX=224308 GN=lytE PE=1 SV=1 |
| P45296 | 7.70e-11 | 244 | 347 | 73 | 172 | Probable endopeptidase NlpC homolog OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=nlpC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
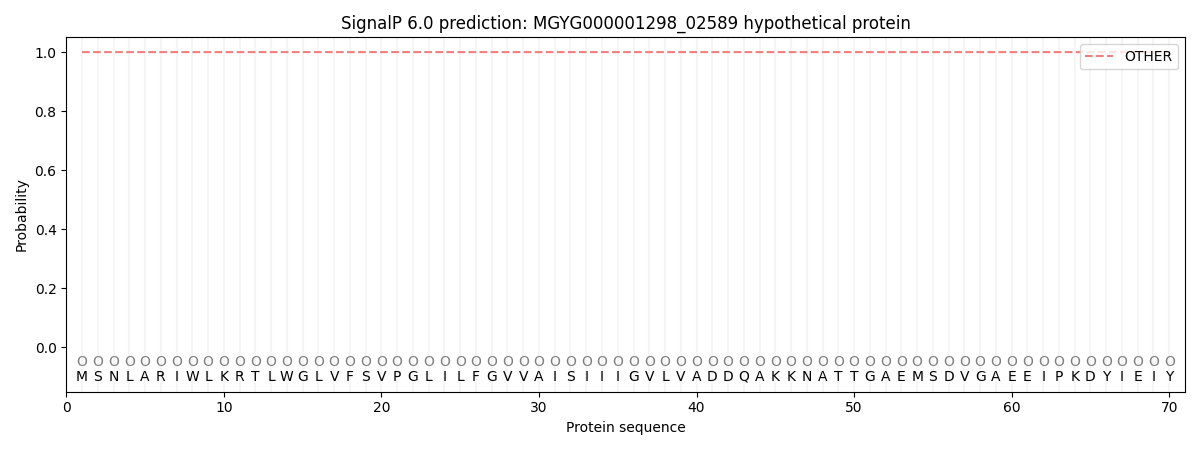
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000039 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |