You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001304_00108
You are here: Home > Sequence: MGYG000001304_00108
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Erysipelatoclostridium spiroforme | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Erysipelatoclostridium; Erysipelatoclostridium spiroforme | |||||||||||
| CAZyme ID | MGYG000001304_00108 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 128486; End: 130600 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 191 | 399 | 7.6e-23 | 0.6079295154185022 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 1.13e-26 | 1 | 436 | 1 | 385 | Predicted peptidase [General function prediction only]. |
| pfam18435 | EstA_Ig_like | 1.85e-18 | 25 | 151 | 4 | 116 | Esterase Ig-like N-terminal domain. This is an N-terminal immunoglobulin (Ig)-like domain found in esterases such as EstA. Analysis of the EstA structure confirms that it is a member of the alpha/beta hydrolase family, with a conserved Ser-Asp-His catalytic triad. The Ig-like domain presumably plays a role in the multimerization of EstA into an unusual hexameric structure. Additionally, it may also participate in the catalysis of EstA by guiding the substrate to the active site. |
| TIGR01840 | esterase_phb | 3.21e-16 | 199 | 395 | 4 | 202 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| COG0400 | YpfH | 1.35e-09 | 196 | 396 | 7 | 193 | Predicted esterase [General function prediction only]. |
| COG1506 | DAP2 | 4.15e-09 | 185 | 328 | 369 | 503 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQY28682.1 | 9.44e-43 | 443 | 702 | 789 | 1050 |
| QQV07490.1 | 9.44e-43 | 443 | 702 | 789 | 1050 |
| QMW74199.1 | 5.38e-42 | 443 | 702 | 789 | 1050 |
| QPS12532.1 | 5.38e-42 | 443 | 702 | 789 | 1050 |
| QMW74468.1 | 1.02e-40 | 433 | 701 | 817 | 1091 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 8.67e-21 | 45 | 438 | 19 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 3.29e-11 | 516 | 649 | 1667 | 1808 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
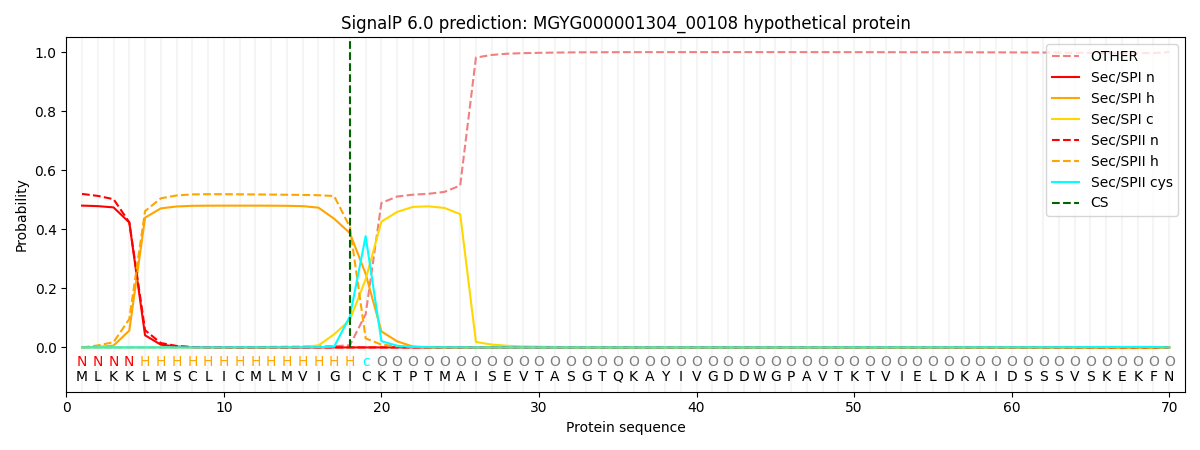
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001563 | 0.466465 | 0.530779 | 0.000501 | 0.000395 | 0.000308 |
