You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001304_00266
You are here: Home > Sequence: MGYG000001304_00266
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
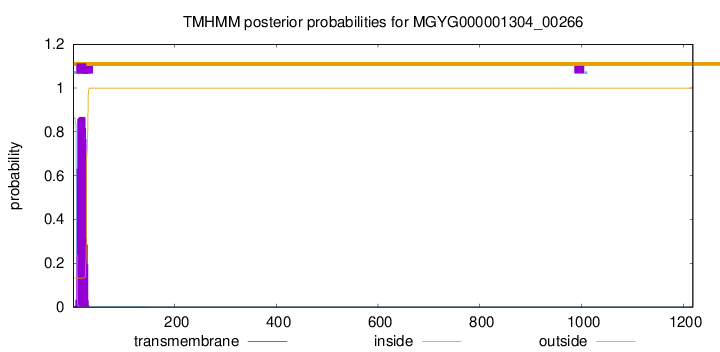
TMHMM annotations
Basic Information help
| Species | Erysipelatoclostridium spiroforme | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Erysipelatoclostridium; Erysipelatoclostridium spiroforme | |||||||||||
| CAZyme ID | MGYG000001304_00266 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 325425; End: 329084 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 5.93e-34 | 681 | 974 | 9 | 323 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG4193 | LytD | 2.77e-17 | 239 | 424 | 53 | 238 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| COG5492 | YjdB | 2.76e-16 | 616 | 909 | 37 | 329 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 3.27e-15 | 920 | 1053 | 26 | 160 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam07538 | ChW | 2.39e-14 | 971 | 1005 | 1 | 35 | Clostridial hydrophobic W. A novel extracellular macromolecular system has been proposed based on the proteins containing ChW repeats. ChW stands for Clostridial hydrophobic with conserved W (tryptophan). This repeat was originally described in Clostridium acetobutylicum but is also found in other Gram-positive bacteria including Enterococcus faecalis, Streptococcus agalactiae and Streptomyces coelicolor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QEK17141.1 | 2.33e-105 | 57 | 532 | 53 | 494 |
| QNM11023.1 | 1.88e-104 | 87 | 535 | 132 | 554 |
| QSI26463.1 | 5.58e-102 | 94 | 543 | 94 | 519 |
| QJA02388.1 | 5.49e-99 | 42 | 548 | 32 | 525 |
| QIX08957.1 | 7.38e-98 | 42 | 503 | 32 | 484 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 3.30e-24 | 130 | 422 | 48 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 1.14e-22 | 130 | 422 | 416 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 1.31e-22 | 130 | 422 | 460 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| P35838 | 2.62e-07 | 828 | 915 | 229 | 318 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
| P33747 | 3.19e-06 | 835 | 913 | 38 | 115 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
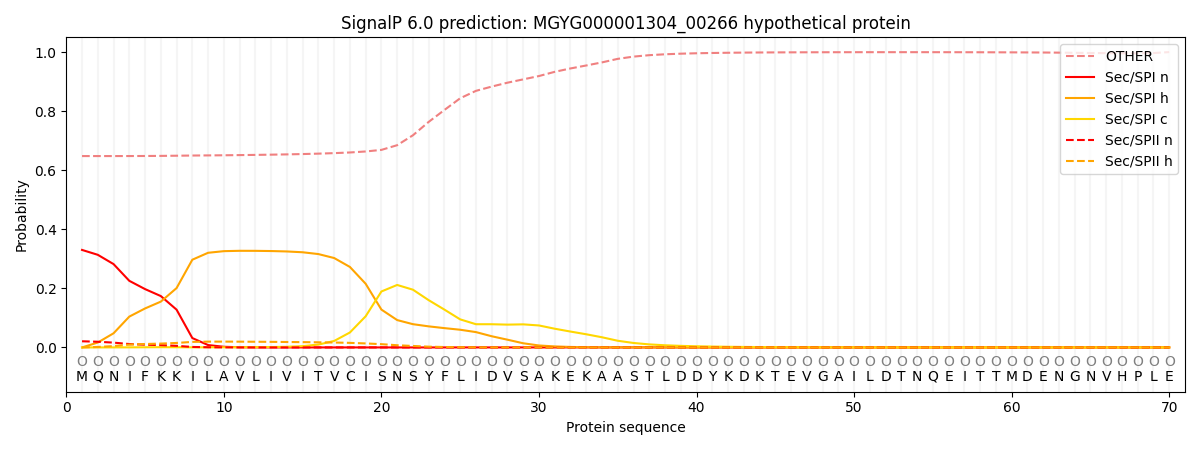
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.662995 | 0.311228 | 0.022620 | 0.000711 | 0.000425 | 0.002013 |