You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001304_01445
You are here: Home > Sequence: MGYG000001304_01445
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
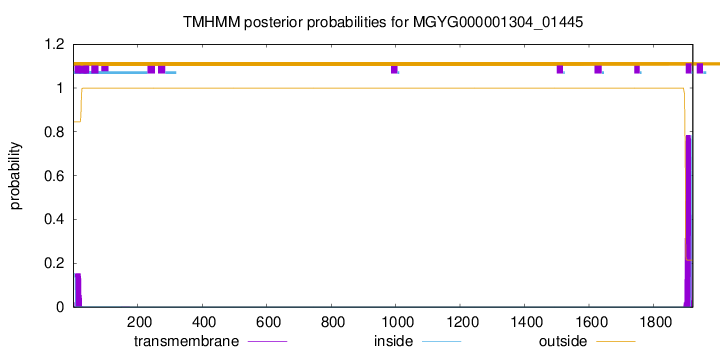
TMHMM annotations
Basic Information help
| Species | Erysipelatoclostridium spiroforme | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelatoclostridiaceae; Erysipelatoclostridium; Erysipelatoclostridium spiroforme | |||||||||||
| CAZyme ID | MGYG000001304_01445 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 133283; End: 139051 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 1407 | 1530 | 4.8e-18 | 0.8951612903225806 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.88e-16 | 1400 | 1531 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 2.99e-06 | 309 | 422 | 15 | 111 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd14791 | GH36 | 5.50e-06 | 658 | 871 | 6 | 194 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| cd00057 | FA58C | 8.78e-06 | 1396 | 1490 | 9 | 95 | Substituted updates: Jan 31, 2002 |
| pfam00754 | F5_F8_type_C | 3.15e-05 | 127 | 254 | 6 | 116 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOY54034.1 | 0.0 | 282 | 1670 | 108 | 1493 |
| ALG48827.1 | 0.0 | 282 | 1641 | 108 | 1463 |
| ABG83191.1 | 0.0 | 282 | 1670 | 108 | 1493 |
| AXH52486.1 | 0.0 | 282 | 1641 | 108 | 1463 |
| ATD48595.1 | 0.0 | 282 | 1670 | 108 | 1493 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2V72_A | 3.83e-10 | 1384 | 1533 | 2 | 141 | Thestructure of the family 32 CBM from C. perfringens NanJ in complex with galactose [Clostridium perfringens] |
| 1W8N_A | 1.99e-09 | 1413 | 1533 | 477 | 597 | Contributionof the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens. [Micromonospora viridifaciens],1W8O_A Contribution of the Active Site Aspartic Acid to Catalysis in the Bacterial Neuraminidase from Micromonospora viridifaciens [Micromonospora viridifaciens] |
| 1WCQ_A | 1.99e-09 | 1413 | 1533 | 477 | 597 | Mutagenesisof the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_B Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens],1WCQ_C Mutagenesis of the Nucleophilic Tyrosine in a Bacterial Sialidase to Phenylalanine. [Micromonospora viridifaciens] |
| 2BER_A | 1.99e-09 | 1413 | 1533 | 477 | 597 | Y370GActive Site Mutant of the Sialidase from Micromonospora viridifaciens in complex with beta-Neu5Ac (sialic acid). [Micromonospora viridifaciens] |
| 2BZD_A | 1.99e-09 | 1413 | 1533 | 477 | 597 | Galactoserecognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_B Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens],2BZD_C Galactose recognition by the carbohydrate-binding module of a bacterial sialidase. [Micromonospora viridifaciens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR4 | 1.90e-09 | 1357 | 1533 | 466 | 643 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| E8MGH9 | 1.94e-09 | 1746 | 1876 | 1667 | 1804 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
| Q02834 | 1.15e-08 | 1413 | 1533 | 523 | 643 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| P26831 | 8.17e-08 | 1708 | 1872 | 1389 | 1565 | Hyaluronoglucosaminidase OS=Clostridium perfringens (strain 13 / Type A) OX=195102 GN=nagH PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000340 | 0.998851 | 0.000291 | 0.000188 | 0.000169 | 0.000161 |