You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001307_03518
You are here: Home > Sequence: MGYG000001307_03518
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Providencia stuartii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Enterobacteriaceae; Providencia; Providencia stuartii | |||||||||||
| CAZyme ID | MGYG000001307_03518 | |||||||||||
| CAZy Family | GH20 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2186750; End: 2189149 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH20 | 157 | 587 | 7.1e-118 | 0.973293768545994 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06570 | GH20_chitobiase-like_1 | 3.56e-174 | 162 | 600 | 1 | 311 | A functionally uncharacterized subgroup of the Glycosyl hydrolase family 20 (GH20) catalytic domain found in proteins similar to the chitobiase of Serratia marcescens, a beta-N-1,4-acetylhexosaminidase that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This subgroup lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| cd06563 | GH20_chitobiase-like | 8.17e-129 | 162 | 600 | 1 | 357 | The chitobiase of Serratia marcescens is a beta-N-1,4-acetylhexosaminidase with a glycosyl hydrolase family 20 (GH20) domain that hydrolyzes the beta-1,4-glycosidic linkages in oligomers derived from chitin. Chitin is degraded by a two step process: i) a chitinase hydrolyzes the chitin to oligosaccharides and disaccharides such as di-N-acetyl-D-glucosamine and chitobiose, ii) chitobiase then further degrades these oligomers into monomers. This GH20 domain family includes an N-acetylglucosamidase (GlcNAcase A) from Pseudoalteromonas piscicida and an N-acetylhexosaminidase (SpHex) from Streptomyces plicatus. SpHex lacks the C-terminal PKD (polycystic kidney disease I)-like domain found in the chitobiases. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| pfam00728 | Glyco_hydro_20 | 2.95e-118 | 162 | 587 | 1 | 345 | Glycosyl hydrolase family 20, catalytic domain. This domain has a TIM barrel fold. |
| cd06562 | GH20_HexA_HexB-like | 5.56e-106 | 162 | 602 | 1 | 339 | Beta-N-acetylhexosaminidases catalyze the removal of beta-1,4-linked N-acetyl-D-hexosamine residues from the non-reducing ends of N-acetyl-beta-D-hexosaminides including N-acetylglucosides and N-acetylgalactosides. The hexA and hexB genes encode the alpha- and beta-subunits of the two major beta-N-acetylhexosaminidase isoenzymes, N-acetyl-beta-D-hexosaminidase A (HexA) and beta-N-acetylhexosaminidase B (HexB). Both the alpha and the beta catalytic subunits have a TIM-barrel fold and belong to the glycosyl hydrolase family 20 (GH20). The HexA enzyme is a heterodimer containing one alpha and one beta subunit while the HexB enzyme is a homodimer containing two beta-subunits. Hexosaminidase mutations cause an inability to properly hydrolyze certain sphingolipids which accumulate in lysosomes within the brain, resulting in the lipid storage disorders Tay-Sachs and Sandhoff. Mutations in the alpha subunit cause in a deficiency in the HexA enzyme and result in Tay-Sachs, mutations in the beta-subunit cause in a deficiency in both HexA and HexB enzymes and result in Sandhoff disease. In both disorders GM(2) gangliosides accumulate in lysosomes. The GH20 hexosaminidases are thought to act via a catalytic mechanism in which the catalytic nucleophile is not provided by solvent or the enzyme, but by the substrate itself. |
| COG3525 | Chb | 1.41e-86 | 30 | 415 | 136 | 542 | N-acetyl-beta-hexosaminidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AMG68604.1 | 0.0 | 1 | 799 | 1 | 799 |
| QPN41565.1 | 0.0 | 1 | 799 | 1 | 799 |
| QIB29237.1 | 0.0 | 1 | 799 | 1 | 799 |
| AVE40708.1 | 0.0 | 1 | 799 | 1 | 799 |
| QUC24808.1 | 0.0 | 1 | 799 | 1 | 799 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YHH_A | 2.56e-113 | 27 | 791 | 6 | 672 | X-rayStructure of Flavobacterium johnsoniae chitobiase (FjGH20) [Flavobacterium johnsoniae UW101],6YHH_B X-ray Structure of Flavobacterium johnsoniae chitobiase (FjGH20) [Flavobacterium johnsoniae UW101] |
| 6Q63_A | 4.85e-69 | 4 | 408 | 6 | 429 | BT0459[Bacteroides thetaiotaomicron],6Q63_B BT0459 [Bacteroides thetaiotaomicron],6Q63_C BT0459 [Bacteroides thetaiotaomicron] |
| 7CBN_A | 3.06e-68 | 110 | 601 | 78 | 497 | Crystalstructure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila [Akkermansia muciniphila ATCC BAA-835],7CBO_A Crystal structure of beta-N-acetylhexosaminidase Am0868 from Akkermansia muciniphila in complex with GlcNAc [Akkermansia muciniphila ATCC BAA-835] |
| 1NOU_A | 5.00e-65 | 61 | 600 | 45 | 480 | Nativehuman lysosomal beta-hexosaminidase isoform B [Homo sapiens],1NOU_B Native human lysosomal beta-hexosaminidase isoform B [Homo sapiens],1NOW_A Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) [Homo sapiens],1NOW_B Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) [Homo sapiens],2GJX_B Crystallographic structure of human beta-Hexosaminidase A [Homo sapiens],2GJX_C Crystallographic structure of human beta-Hexosaminidase A [Homo sapiens],2GJX_F Crystallographic structure of human beta-Hexosaminidase A [Homo sapiens],2GJX_G Crystallographic structure of human beta-Hexosaminidase A [Homo sapiens] |
| 1O7A_A | 6.02e-65 | 61 | 600 | 53 | 488 | Humanbeta-Hexosaminidase B [Homo sapiens],1O7A_B Human beta-Hexosaminidase B [Homo sapiens],1O7A_C Human beta-Hexosaminidase B [Homo sapiens],1O7A_D Human beta-Hexosaminidase B [Homo sapiens],1O7A_E Human beta-Hexosaminidase B [Homo sapiens],1O7A_F Human beta-Hexosaminidase B [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49008 | 2.32e-70 | 30 | 429 | 37 | 442 | Beta-hexosaminidase OS=Porphyromonas gingivalis (strain ATCC BAA-308 / W83) OX=242619 GN=nahA PE=3 SV=2 |
| P96155 | 8.96e-69 | 22 | 415 | 133 | 535 | Beta-hexosaminidase OS=Vibrio furnissii OX=29494 GN=exoI PE=1 SV=1 |
| B2UQG6 | 5.88e-68 | 1 | 601 | 1 | 516 | Beta-hexosaminidase Amuc_0868 OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=Amuc_0868 PE=1 SV=1 |
| Q641X3 | 1.68e-64 | 30 | 410 | 23 | 423 | Beta-hexosaminidase subunit alpha OS=Rattus norvegicus OX=10116 GN=Hexa PE=2 SV=1 |
| P07686 | 8.28e-64 | 61 | 600 | 94 | 529 | Beta-hexosaminidase subunit beta OS=Homo sapiens OX=9606 GN=HEXB PE=1 SV=4 |
SignalP and Lipop Annotations help
This protein is predicted as SP
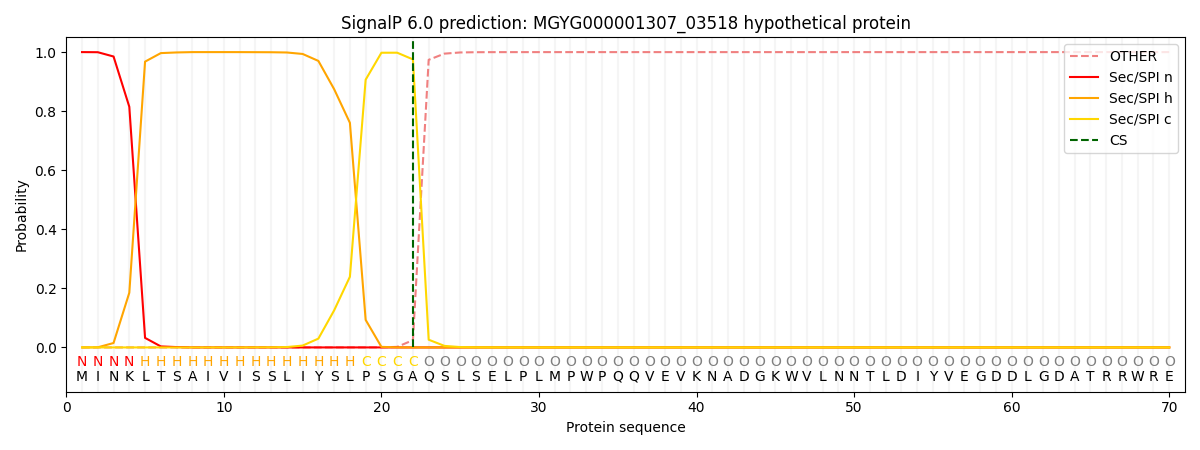
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.999002 | 0.000196 | 0.000183 | 0.000170 | 0.000145 |
