You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001310_01417
You are here: Home > Sequence: MGYG000001310_01417
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
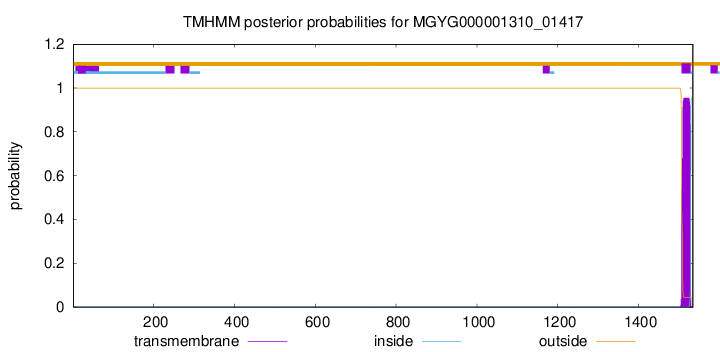
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter lactaris | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter lactaris | |||||||||||
| CAZyme ID | MGYG000001310_01417 | |||||||||||
| CAZy Family | GH101 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2011; End: 6618 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH101 | 334 | 872 | 9.4e-92 | 0.8062234794908062 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14244 | GH_101_like | 2.28e-80 | 564 | 862 | 1 | 298 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam12905 | Glyco_hydro_101 | 2.11e-69 | 551 | 806 | 1 | 268 | Endo-alpha-N-acetylgalactosaminidase. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by the S. pneumoniae protein Endo-alpha-N-acetylgalactosaminidase, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. |
| pfam18080 | Gal_mutarotas_3 | 1.58e-46 | 331 | 549 | 1 | 242 | Galactose mutarotase-like fold domain. This domain is found in endo-alpha-N-acetylgalactosaminidase present in Streptococcus pneumoniae. Endo-alpha-N-acetylgalactosaminidase is a cell surface-anchored glycoside hydrolase involved in the breakdown of mucin type O-linked glycans. The domain, known as domain 2, exhibits strong structural similarlity to the galactose mutarotase-like fold but lacks the active site residues. Domains, found in a number of glycoside hydrolases, structurally similar to domain 2 confer stability to the multidomain architectures. |
| pfam17451 | Glyco_hyd_101C | 3.50e-23 | 845 | 946 | 1 | 110 | Glycosyl hydrolase 101 beta sandwich domain. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae is largely determined by the ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. This family is the enzymatic region, EC:3.2.1.97, of the cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins. This reaction is exemplified by a S. pneumoniae protein, where Asp764 is the catalytic nucleophile-base and Glu796 the catalytic proton donor. This domain represents C-terminal the beta sandwich domain. |
| pfam03442 | CBM_X2 | 4.70e-16 | 50 | 132 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBK22105.1 | 0.0 | 2 | 1389 | 177 | 1565 |
| BBK62189.1 | 0.0 | 2 | 1389 | 177 | 1565 |
| ATD58436.1 | 0.0 | 13 | 1406 | 371 | 1740 |
| ATD54117.1 | 0.0 | 13 | 1406 | 371 | 1740 |
| QBJ76357.1 | 0.0 | 13 | 1406 | 371 | 1740 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5A55_A | 1.63e-93 | 330 | 1240 | 20 | 1019 | Thenative structure of GH101 from Streptococcus pneumoniae TIGR4 [Streptococcus pneumoniae TIGR4] |
| 5A57_A | 1.76e-93 | 330 | 1240 | 18 | 1017 | Thestructure of GH101 from Streptococcus pneumoniae TIGR4 in complex with PUGT [Streptococcus pneumoniae TIGR4] |
| 5A56_A | 1.76e-93 | 330 | 1240 | 18 | 1017 | Thestructure of GH101 from Streptococcus pneumoniae TIGR4 in complex with 1-O-methyl-T-antigen [Streptococcus pneumoniae TIGR4] |
| 5A59_A | 4.33e-93 | 330 | 1240 | 18 | 1017 | Thestructure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with T-antigen [Streptococcus pneumoniae TIGR4],5A5A_A The structure of GH101 E796Q mutant from Streptococcus pneumoniae TIGR4 in complex with PNP-T-antigen [Streptococcus pneumoniae TIGR4] |
| 5A58_A | 7.89e-93 | 330 | 1240 | 18 | 1017 | Thestructure of GH101 D764N mutant from Streptococcus pneumoniae TIGR4 in complex with serinyl T-antigen [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q2MGH6 | 1.20e-91 | 297 | 1240 | 299 | 1332 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=SP_0368 PE=1 SV=1 |
| Q8DR60 | 1.60e-91 | 297 | 1240 | 299 | 1332 | Endo-alpha-N-acetylgalactosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=spr0328 PE=1 SV=1 |
| A9WNA0 | 1.83e-87 | 329 | 1238 | 50 | 973 | Putative endo-alpha-N-acetylgalactosaminidase OS=Renibacterium salmoninarum (strain ATCC 33209 / DSM 20767 / JCM 11484 / NBRC 15589 / NCIMB 2235) OX=288705 GN=RSal33209_1326 PE=3 SV=2 |
| Q3MUH7 | 7.34e-06 | 50 | 125 | 783 | 858 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
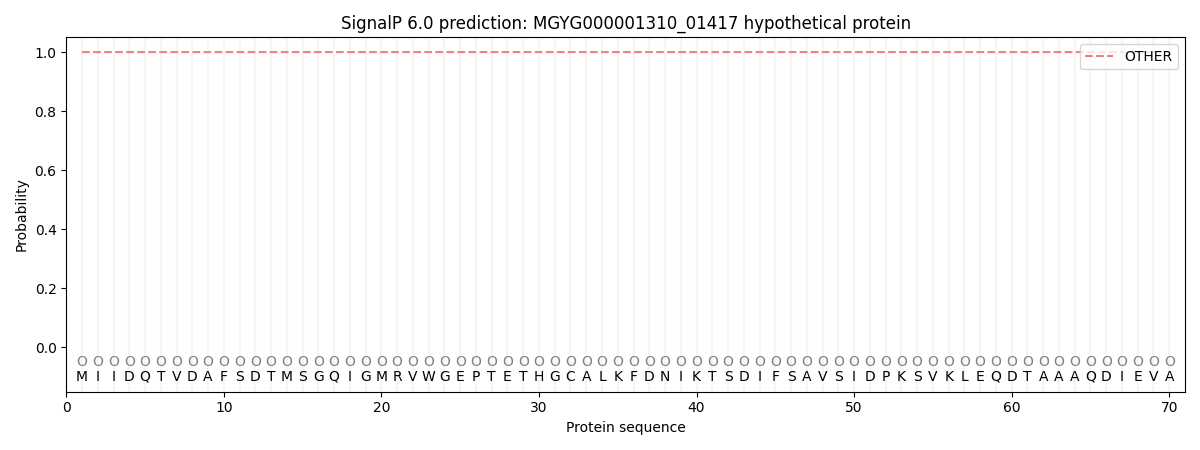
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000038 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |