You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001312_00387
You are here: Home > Sequence: MGYG000001312_00387
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Helicobacter_D pullorum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Helicobacteraceae; Helicobacter_D; Helicobacter_D pullorum | |||||||||||
| CAZyme ID | MGYG000001312_00387 | |||||||||||
| CAZy Family | GT66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 75784; End: 76992 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT66 | 3 | 281 | 7.3e-31 | 0.4069264069264069 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEJ08217.1 | 9.50e-281 | 1 | 402 | 292 | 693 |
| ADM36001.1 | 7.35e-278 | 1 | 402 | 292 | 693 |
| QOQ98180.1 | 8.23e-84 | 8 | 401 | 335 | 730 |
| CAE09214.1 | 7.65e-28 | 17 | 325 | 282 | 592 |
| VEG81426.1 | 7.65e-28 | 17 | 325 | 282 | 592 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3RCE_A | 7.12e-15 | 17 | 323 | 281 | 595 | Bacterialoligosaccharyltransferase PglB [Campylobacter lari] |
| 5OGL_A | 2.96e-14 | 17 | 323 | 281 | 595 | Structureof bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog [Campylobacter lari RM2100] |
| 6GXC_A | 2.98e-14 | 17 | 323 | 281 | 595 | Bacterialoligosaccharyltransferase PglB in complex with an inhibitory peptide and a reactive lipid-linked oligosaccharide analog [Campylobacter lari RM2100] |
| 3AAG_A | 2.89e-11 | 184 | 315 | 28 | 164 | Crystalstructure of C. jejuni pglb C-terminal domain [Campylobacter jejuni RM1221],3AAG_B Crystal structure of C. jejuni pglb C-terminal domain [Campylobacter jejuni RM1221] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5HTX9 | 1.32e-22 | 20 | 315 | 282 | 586 | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter jejuni (strain RM1221) OX=195099 GN=pglB PE=1 SV=1 |
| Q0P9C8 | 1.78e-22 | 20 | 315 | 282 | 586 | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=pglB PE=1 SV=1 |
| B9KDD4 | 2.90e-15 | 17 | 323 | 281 | 595 | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter lari (strain RM2100 / D67 / ATCC BAA-1060) OX=306263 GN=pglB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
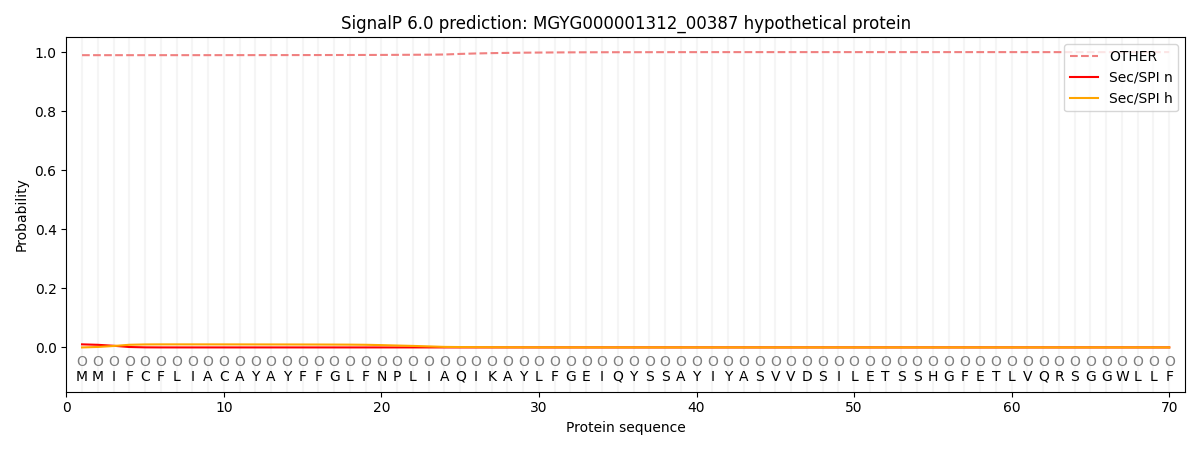
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.990204 | 0.009710 | 0.000026 | 0.000015 | 0.000014 | 0.000025 |
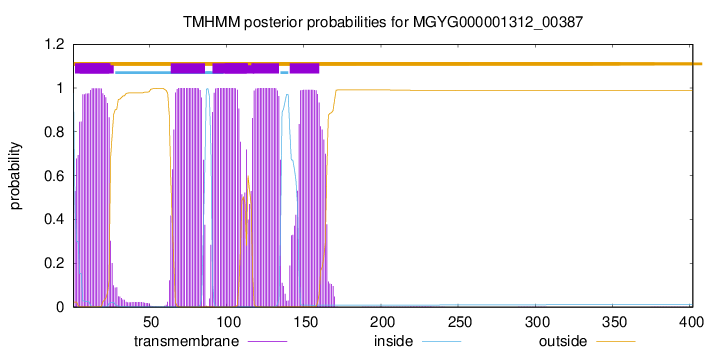
TMHMM Annotations download full data without filtering help
| start | end |
|---|---|
| 2 | 24 |
| 64 | 86 |
| 91 | 113 |
| 117 | 134 |
| 141 | 160 |
