You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001313_00402
You are here: Home > Sequence: MGYG000001313_00402
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides eggerthii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides eggerthii | |||||||||||
| CAZyme ID | MGYG000001313_00402 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 432165; End: 433721 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 169 | 511 | 6e-53 | 0.554364471669219 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT44995.1 | 0.0 | 1 | 518 | 1 | 518 |
| QRQ49249.1 | 0.0 | 1 | 518 | 1 | 518 |
| QDO68502.1 | 7.25e-287 | 1 | 517 | 1 | 515 |
| QDH57425.1 | 4.39e-281 | 1 | 517 | 1 | 515 |
| QGT71933.1 | 2.54e-233 | 3 | 517 | 1 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T3B_A | 1.23e-143 | 51 | 517 | 24 | 480 | ChainA, Glycoside Hydrolase [Phocaeicola plebeius],5T3B_B Chain B, Glycoside Hydrolase [Phocaeicola plebeius] |
| 4BQ2_A | 4.56e-13 | 105 | 510 | 233 | 743 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 1.05e-12 | 105 | 510 | 233 | 743 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
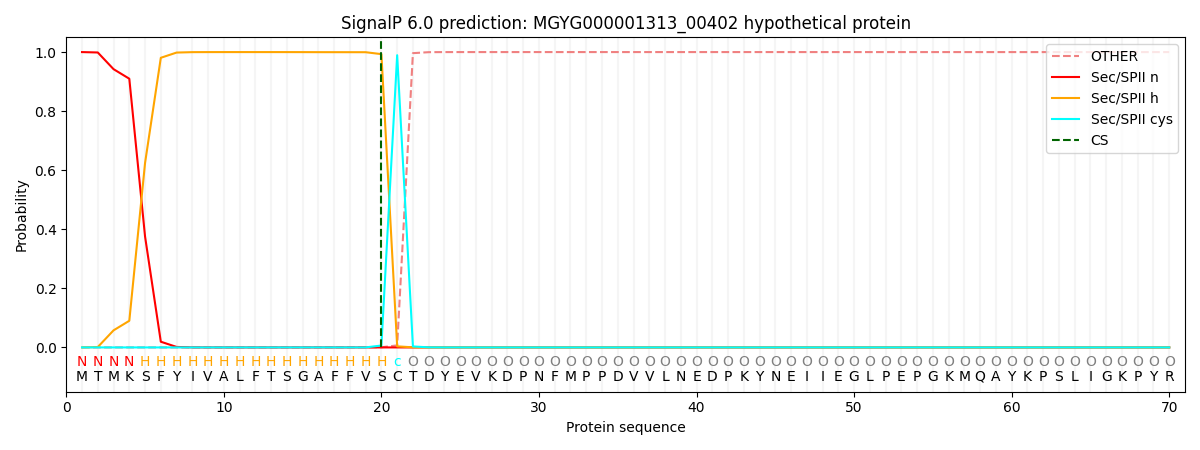
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000010 | 1.000058 | 0.000000 | 0.000000 | 0.000000 |
