You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001313_00820
You are here: Home > Sequence: MGYG000001313_00820
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
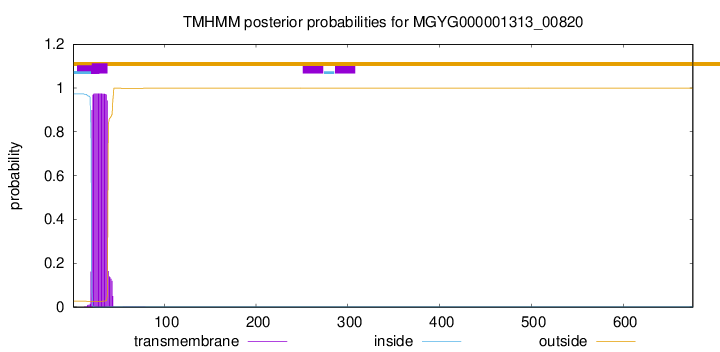
TMHMM annotations
Basic Information help
| Species | Bacteroides eggerthii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides eggerthii | |||||||||||
| CAZyme ID | MGYG000001313_00820 | |||||||||||
| CAZy Family | GH120 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 975690; End: 977720 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH120 | 336 | 426 | 4e-31 | 0.989010989010989 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 1.19e-06 | 279 | 445 | 13 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 2.96e-04 | 335 | 453 | 32 | 140 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam07602 | DUF1565 | 3.79e-04 | 46 | 84 | 2 | 40 | Protein of unknown function (DUF1565). These proteins share a region of homology in their N termini, and are found in several phylogenetically diverse bacteria and in the archaeon Methanosarcina acetivorans. Some of these proteins also contain characterized domains such as pfam00395 and pfam03422. |
| pfam13229 | Beta_helix | 0.009 | 332 | 470 | 6 | 142 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ48868.1 | 0.0 | 1 | 676 | 1 | 676 |
| QUT44521.1 | 0.0 | 1 | 676 | 1 | 676 |
| AUC24119.1 | 2.39e-220 | 38 | 670 | 3 | 634 |
| QWX84993.1 | 1.61e-219 | 30 | 670 | 12 | 649 |
| QNO17287.1 | 4.78e-207 | 39 | 671 | 2 | 646 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VST_A | 8.35e-194 | 40 | 670 | 2 | 638 | Thecomplex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_B The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_C The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_D The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_A The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_B The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_C The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_D The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_A The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_B The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_C The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_D The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
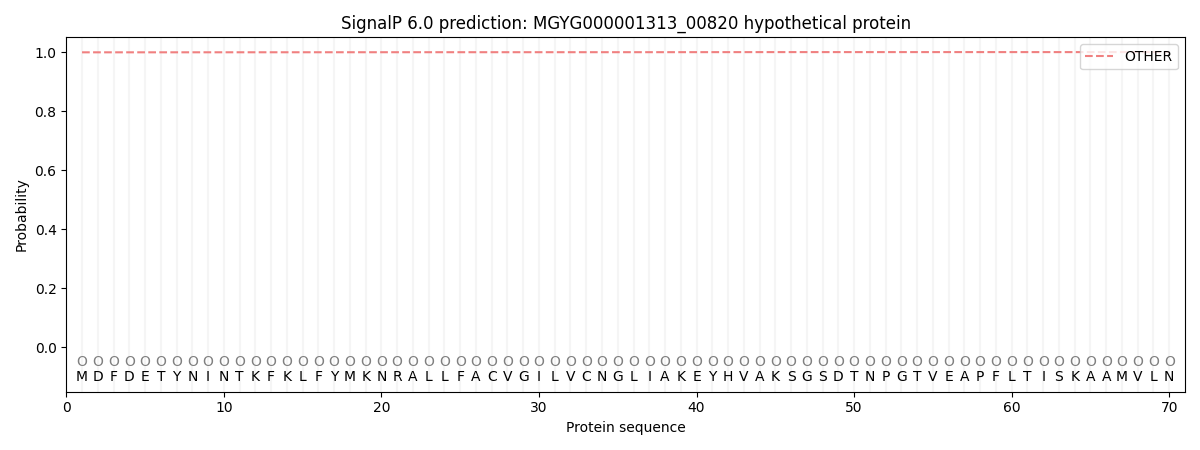
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999295 | 0.000487 | 0.000154 | 0.000002 | 0.000001 | 0.000076 |