You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001313_01273
You are here: Home > Sequence: MGYG000001313_01273
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides eggerthii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides eggerthii | |||||||||||
| CAZyme ID | MGYG000001313_01273 | |||||||||||
| CAZy Family | GH165 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 562837; End: 564501 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH165 | 17 | 553 | 2.9e-223 | 0.7921259842519685 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08984 | GH43-like | 9.45e-05 | 121 | 223 | 142 | 217 | Glycosyl hydrolase family 43. This glycosyl hydrolase family 43 (GH43)-like subfamily includes uncharacterized enzymes similar to those with beta-1,4-xylosidase (xylan 1,4-beta-xylosidase; EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanase and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. These are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many of the enzymes in this family display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRQ47328.1 | 0.0 | 1 | 554 | 1 | 554 |
| AVM55299.1 | 0.0 | 1 | 552 | 1 | 552 |
| QGS17344.1 | 0.0 | 17 | 553 | 16 | 552 |
| ATA82561.1 | 0.0 | 17 | 553 | 16 | 552 |
| QUT46502.1 | 2.79e-305 | 22 | 552 | 27 | 559 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
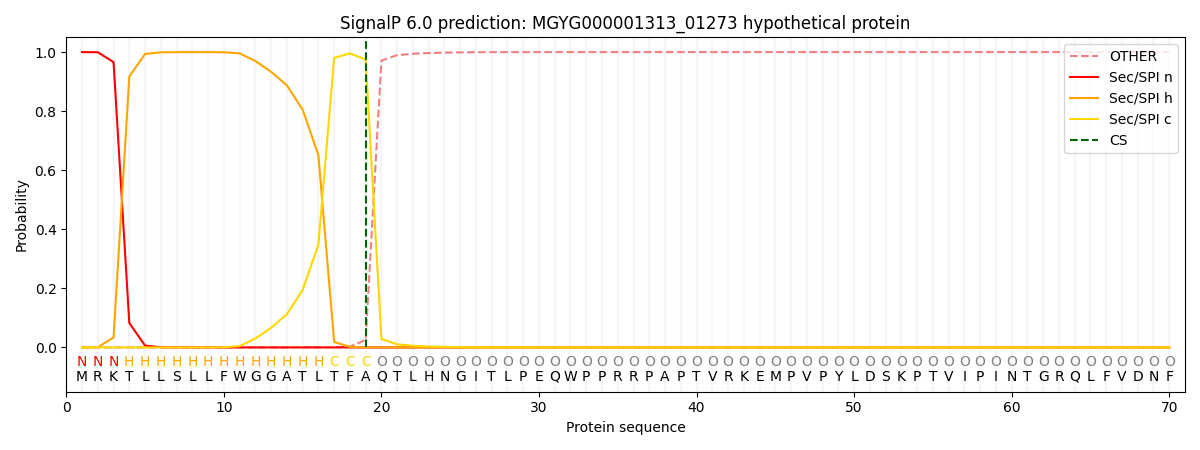
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000481 | 0.998670 | 0.000272 | 0.000197 | 0.000189 | 0.000176 |
