You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001314_00547
You are here: Home > Sequence: MGYG000001314_00547
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
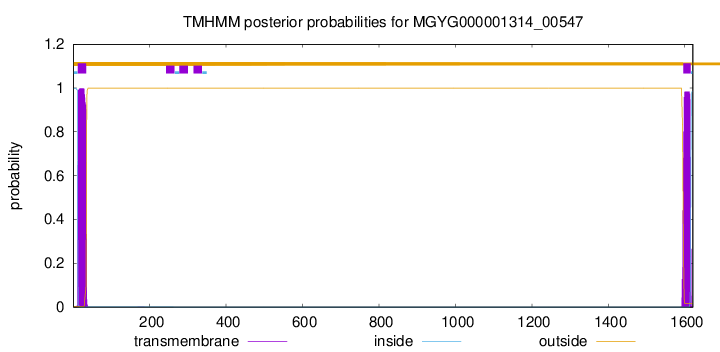
TMHMM annotations
Basic Information help
| Species | Bacteroides_F pectinophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Bacteroides_F; Bacteroides_F pectinophilus | |||||||||||
| CAZyme ID | MGYG000001314_00547 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 66133; End: 70998 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 934 | 1319 | 7.1e-41 | 0.9270833333333334 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.06e-18 | 923 | 1313 | 79 | 386 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 7.94e-18 | 1041 | 1333 | 68 | 296 | Pectinesterase. |
| PLN02773 | PLN02773 | 1.66e-14 | 1050 | 1259 | 90 | 239 | pectinesterase |
| PLN02301 | PLN02301 | 7.84e-14 | 1054 | 1325 | 315 | 535 | pectinesterase/pectinesterase inhibitor |
| PLN03043 | PLN03043 | 9.08e-13 | 1119 | 1311 | 329 | 500 | Probable pectinesterase/pectinesterase inhibitor; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFK85374.1 | 1.39e-265 | 360 | 1345 | 749 | 1731 |
| ADL35206.1 | 9.21e-214 | 346 | 1338 | 118 | 1123 |
| QBZ98893.1 | 4.78e-195 | 254 | 1338 | 63 | 1133 |
| AOW17789.1 | 8.61e-194 | 255 | 1340 | 461 | 1524 |
| AVR45655.1 | 1.02e-190 | 224 | 1338 | 298 | 1397 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
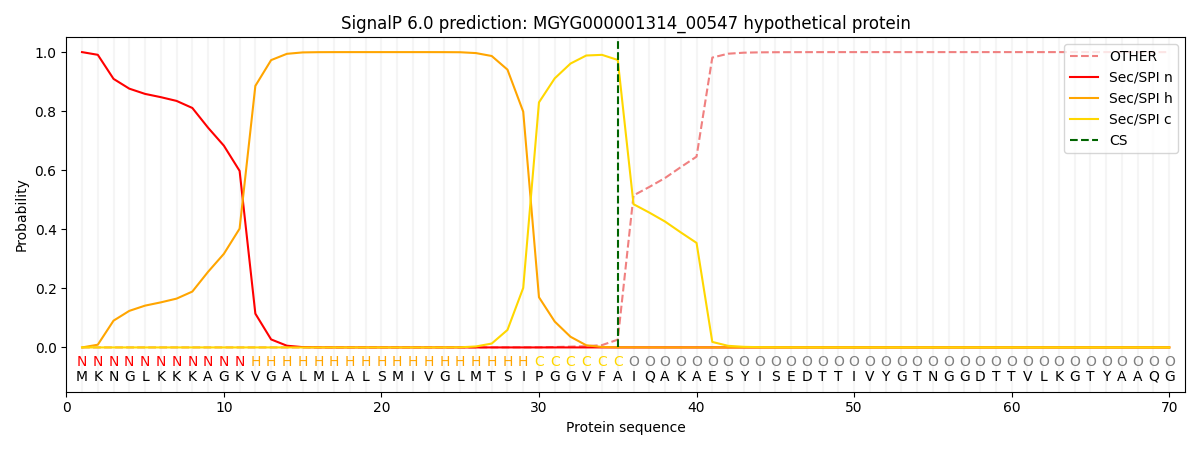
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000351 | 0.998970 | 0.000203 | 0.000178 | 0.000146 | 0.000137 |