You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001314_02453
You are here: Home > Sequence: MGYG000001314_02453
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
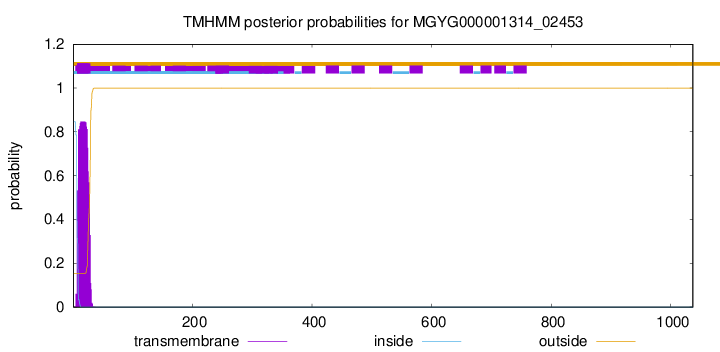
TMHMM annotations
Basic Information help
| Species | Bacteroides_F pectinophilus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Bacteroides_F; Bacteroides_F pectinophilus | |||||||||||
| CAZyme ID | MGYG000001314_02453 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1930545; End: 1933658 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 225 | 391 | 1e-45 | 0.801980198019802 |
| CBM77 | 606 | 712 | 1.2e-33 | 0.9902912621359223 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 1.15e-55 | 96 | 496 | 2 | 340 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam18283 | CBM77 | 3.40e-34 | 604 | 714 | 1 | 108 | Carbohydrate binding module 77. This domain is the non-catalytic carbohydrate binding module 77 (CBM77) present in Ruminococcus flavefaciens. CBMs fulfil a critical targeting function in plant cell wall depolymerisation. In CBM77, a cluster of conserved basic residues (Lys1092, Lys1107 and Lys1162) confer calcium-independent recognition of homogalacturonan. |
| smart00656 | Amb_all | 1.06e-31 | 227 | 393 | 19 | 190 | Amb_all domain. |
| pfam00544 | Pec_lyase_C | 2.97e-20 | 227 | 389 | 38 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| NF033930 | pneumo_PspA | 1.80e-13 | 722 | 1006 | 408 | 659 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU23076.1 | 9.36e-226 | 22 | 722 | 33 | 711 |
| ADL35105.1 | 8.06e-221 | 40 | 726 | 61 | 754 |
| AWK03841.1 | 2.61e-188 | 36 | 727 | 21 | 749 |
| AYN05640.1 | 2.08e-181 | 33 | 715 | 19 | 762 |
| QHJ07965.1 | 1.75e-180 | 30 | 715 | 15 | 667 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FU5_A | 1.81e-25 | 604 | 716 | 6 | 113 | Thecomplexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition [Ruminococcus flavefaciens] |
| 3VMV_A | 4.40e-23 | 181 | 463 | 31 | 321 | Crystalstructure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
| 1AIR_A | 1.71e-17 | 189 | 404 | 43 | 272 | ChainA, PECTATE LYASE C [Dickeya chrysanthemi],1O88_A Chain A, Pectate Lyase C [Dickeya chrysanthemi],1O8D_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8E_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8F_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8G_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8H_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8I_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8J_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8K_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8L_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1O8M_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi],1PLU_A Chain A, Protein (pectate Lyase C) [Dickeya chrysanthemi],2PEC_A Chain A, PECTATE LYASE C [Dickeya chrysanthemi] |
| 2EWE_A | 4.13e-17 | 189 | 404 | 43 | 272 | ChainA, Pectate lyase C [Dickeya chrysanthemi] |
| 1VBL_A | 5.66e-15 | 227 | 389 | 135 | 330 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1B6T1 | 7.79e-25 | 176 | 398 | 64 | 281 | Pectate trisaccharide-lyase OS=Bacillus sp. OX=1409 GN=pel PE=1 SV=1 |
| Q65DC2 | 7.79e-25 | 176 | 398 | 64 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=BLi04129 PE=3 SV=1 |
| Q8GCB2 | 7.79e-25 | 176 | 398 | 64 | 281 | Pectate trisaccharide-lyase OS=Bacillus licheniformis OX=1402 GN=pelA PE=1 SV=1 |
| P0C1C2 | 1.10e-18 | 189 | 404 | 64 | 293 | Pectate lyase 3 OS=Pectobacterium carotovorum OX=554 GN=pel3 PE=1 SV=1 |
| P0C1C3 | 1.48e-18 | 189 | 404 | 64 | 293 | Pectate lyase 3 OS=Pectobacterium carotovorum subsp. carotovorum OX=555 GN=pel3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
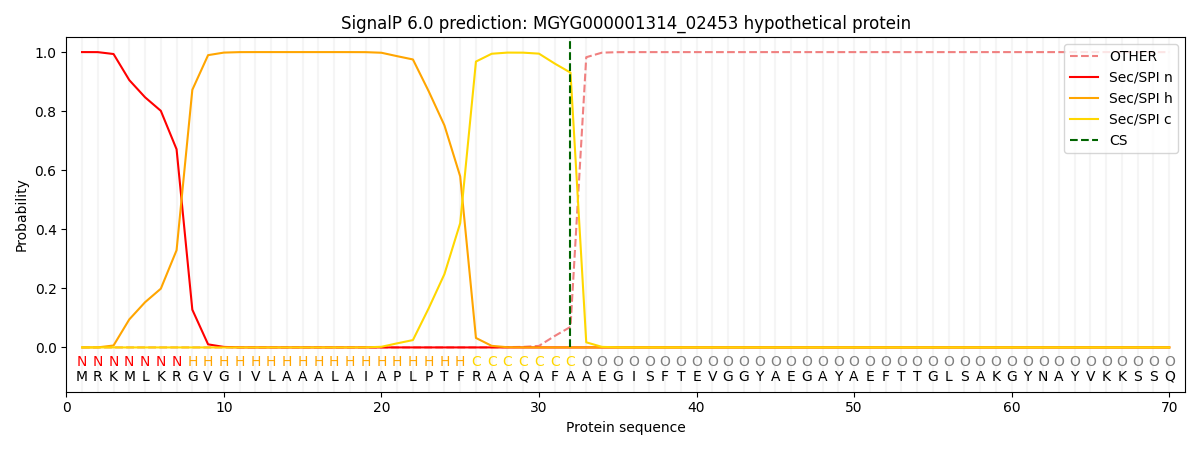
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000269 | 0.999015 | 0.000180 | 0.000213 | 0.000172 | 0.000146 |