You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001315_02662
You are here: Home > Sequence: MGYG000001315_02662
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
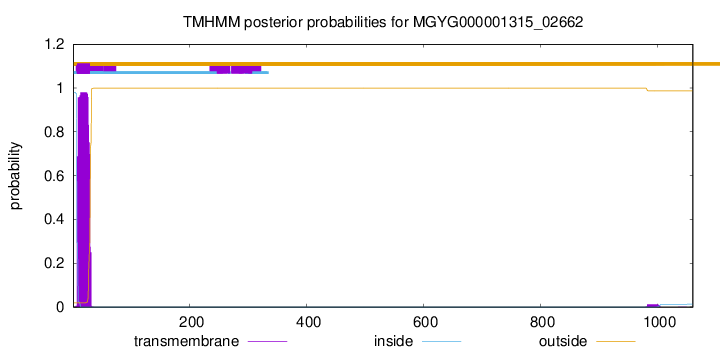
TMHMM annotations
Basic Information help
| Species | Bariatricus comes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Bariatricus; Bariatricus comes | |||||||||||
| CAZyme ID | MGYG000001315_02662 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 274474; End: 277659 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 285 | 438 | 1.4e-22 | 0.7659574468085106 |
| CBM13 | 637 | 772 | 6.4e-20 | 0.6914893617021277 |
| CBM13 | 774 | 866 | 1.2e-17 | 0.4734042553191489 |
| CBM13 | 439 | 589 | 1.7e-16 | 0.7553191489361702 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13529 | Peptidase_C39_2 | 4.31e-29 | 877 | 1034 | 1 | 139 | Peptidase_C39 like family. |
| pfam14200 | RicinB_lectin_2 | 9.80e-24 | 764 | 852 | 2 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.28e-19 | 712 | 804 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| COG4990 | YvpB | 8.74e-18 | 865 | 1060 | 10 | 193 | Predicted cysteine peptidase, C39 family [General function prediction only]. |
| pfam14200 | RicinB_lectin_2 | 1.56e-17 | 321 | 410 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRT49575.1 | 0.0 | 1 | 1061 | 1 | 1061 |
| BAK44091.1 | 7.76e-70 | 111 | 869 | 128 | 885 |
| QRT49576.1 | 7.21e-64 | 270 | 784 | 413 | 926 |
| QRT50581.1 | 2.93e-27 | 272 | 784 | 120 | 784 |
| QOL34941.1 | 6.40e-24 | 106 | 391 | 750 | 1036 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3ERV_A | 1.34e-07 | 878 | 1039 | 37 | 200 | Crystalstructure of an putative C39-like peptidase from Bacillus anthracis [Bacillus anthracis] |
| 5B2H_A | 3.80e-06 | 556 | 628 | 210 | 280 | Crystalstructure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum],5B2H_B Crystal structure of HA33 from Clostridium botulinum serotype C strain Yoichi [Clostridium botulinum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34735 | 3.55e-07 | 885 | 1061 | 73 | 249 | Uncharacterized protein YvpB OS=Bacillus subtilis (strain 168) OX=224308 GN=yvpB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
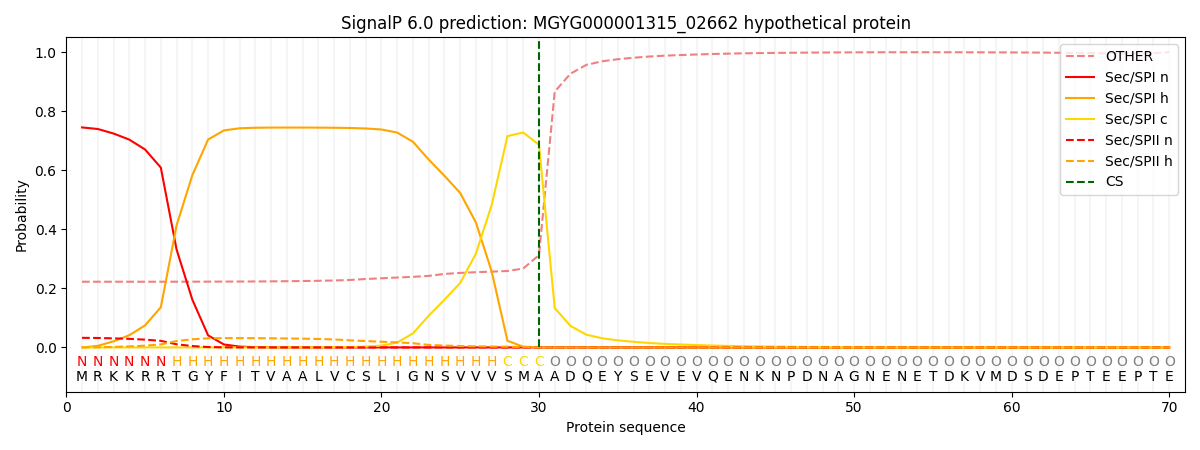
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.241026 | 0.721637 | 0.034730 | 0.001189 | 0.000639 | 0.000749 |