You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001321_00358
You are here: Home > Sequence: MGYG000001321_00358
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
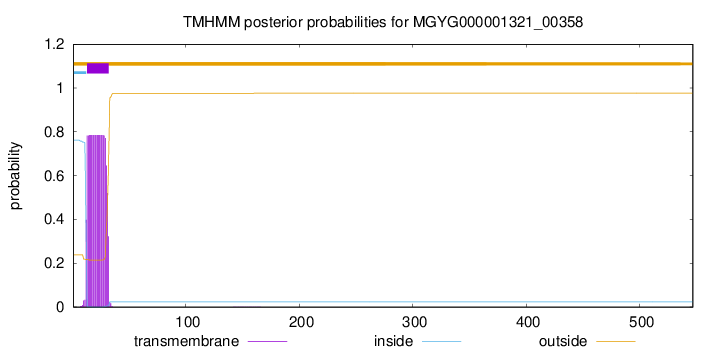
TMHMM annotations
Basic Information help
| Species | Desulfovibrio piger | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Desulfobacterota; Desulfovibrionia; Desulfovibrionales; Desulfovibrionaceae; Desulfovibrio; Desulfovibrio piger | |||||||||||
| CAZyme ID | MGYG000001321_00358 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 224218; End: 225861 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 110 | 251 | 4.6e-29 | 0.8666666666666667 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd16894 | MltD-like | 1.13e-60 | 115 | 250 | 1 | 128 | Membrane-bound lytic murein transglycosylase D and similar proteins. Lytic transglycosylases (LT) catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc). Membrane-bound lytic murein transglycosylase D protein (MltD) family members may have one or more small LysM domains, which may contribute to peptidoglycan binding. Unlike the similar "goose-type" lysozymes, LTs also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| PRK10783 | mltD | 1.29e-55 | 104 | 547 | 101 | 450 | membrane-bound lytic murein transglycosylase D; Provisional |
| pfam01464 | SLT | 2.17e-27 | 116 | 221 | 7 | 113 | Transglycosylase SLT domain. This family is distantly related to pfam00062. Members are found in phages, type II, type III and type IV secretion systems. |
| PRK06347 | PRK06347 | 4.39e-18 | 293 | 545 | 357 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 3.56e-17 | 352 | 545 | 339 | 523 | 1,4-beta-N-acetylmuramoylhydrolase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SFV73355.1 | 0.0 | 1 | 547 | 1 | 547 |
| AMD88750.1 | 1.04e-229 | 17 | 545 | 16 | 550 |
| QTO41906.1 | 1.10e-192 | 12 | 545 | 2 | 563 |
| ATD82166.1 | 7.18e-186 | 7 | 545 | 11 | 553 |
| SPD34874.1 | 7.18e-186 | 7 | 545 | 11 | 553 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PXV_A | 2.04e-07 | 502 | 545 | 4 | 47 | CrystalStructure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],4PXV_B Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],4PXV_C Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],4PXV_D Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],5YLG_A Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],5YLG_B Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],5YLG_C Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis],5YLG_D Crystal Structure of LysM domain from pteris ryukyuensis chitinase A [Pteris ryukyuensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AEZ8 | 3.11e-35 | 81 | 543 | 75 | 440 | Membrane-bound lytic murein transglycosylase D OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=mltD PE=3 SV=1 |
| P0AEZ7 | 3.11e-35 | 81 | 543 | 75 | 440 | Membrane-bound lytic murein transglycosylase D OS=Escherichia coli (strain K12) OX=83333 GN=mltD PE=1 SV=1 |
| P32820 | 2.73e-13 | 101 | 210 | 19 | 129 | Putative tributyltin chloride resistance protein OS=Alteromonas sp. (strain M-1) OX=29457 GN=tbtA PE=3 SV=1 |
| P37710 | 8.50e-12 | 350 | 546 | 571 | 737 | Autolysin OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0799 PE=1 SV=2 |
| P39046 | 1.57e-09 | 350 | 545 | 342 | 530 | Muramidase-2 OS=Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R) OX=768486 GN=EHR_05900 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
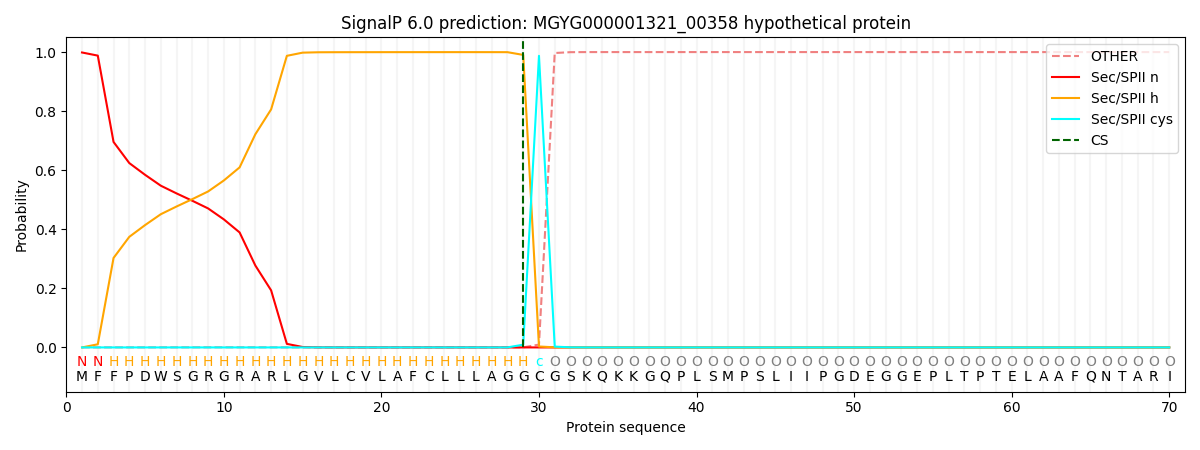
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000021 | 0.000243 | 0.999784 | 0.000006 | 0.000001 | 0.000001 |