You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001324_00155
Basic Information
help
| Species |
Fusobacterium periodonticum_D
|
| Lineage |
Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium; Fusobacterium periodonticum_D
|
| CAZyme ID |
MGYG000001324_00155
|
| CAZy Family |
GT83 |
| CAZyme Description |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001324 |
2472878 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 168648;
End: 170201
Strand: -
|
No EC number prediction in MGYG000001324_00155.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
5 |
421 |
1.2e-56 |
0.7870370370370371 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.54e-39 |
6 |
495 |
7 |
505 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
1.39e-13 |
62 |
225 |
2 |
159 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| PRK13279
|
arnT |
2.38e-10 |
40 |
234 |
40 |
234 |
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase. |
| COG1928
|
PMT1 |
0.005 |
9 |
223 |
28 |
263 |
Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O67601
|
2.95e-10 |
6 |
308 |
2 |
291 |
Uncharacterized protein aq_1704 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1704 PE=3 SV=1 |
This protein is predicted as OTHER
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.998342
|
0.001342
|
0.000074
|
0.000009
|
0.000007
|
0.000226
|
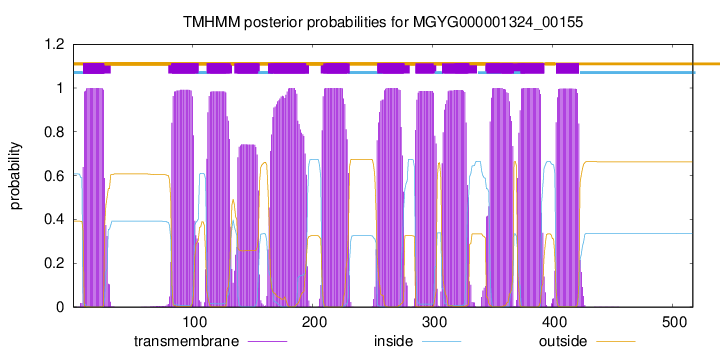
| start |
end |
| 9 |
26 |
| 83 |
105 |
| 112 |
131 |
| 135 |
154 |
| 163 |
194 |
| 209 |
231 |
| 254 |
276 |
| 286 |
301 |
| 308 |
330 |
| 345 |
367 |
| 374 |
393 |
| 403 |
422 |


