You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001330_00580
You are here: Home > Sequence: MGYG000001330_00580
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Helicobacter_D winghamensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Campylobacterota; Campylobacteria; Campylobacterales; Helicobacteraceae; Helicobacter_D; Helicobacter_D winghamensis | |||||||||||
| CAZyme ID | MGYG000001330_00580 | |||||||||||
| CAZy Family | GT66 | |||||||||||
| CAZyme Description | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 153140; End: 155011 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT66 | 16 | 495 | 4.5e-63 | 0.7619047619047619 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1287 | Stt3 | 1.29e-19 | 1 | 479 | 9 | 543 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
| pfam02516 | STT3 | 1.41e-12 | 83 | 459 | 78 | 477 | Oligosaccharyl transferase STT3 subunit. This family consists of the oligosaccharyl transferase STT3 subunit and related proteins. The STT3 subunit is part of the oligosaccharyl transferase (OTase) complex of proteins and is required for its activity. In eukaryotes, OTase transfers a lipid-linked core-oligosaccharide to selected asparagine residues in the ER. In the archaea STT3 occurs alone, rather than in an OTase complex, and is required for N-glycosylation of asparagines. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOQ98472.1 | 0.0 | 1 | 623 | 1 | 618 |
| ADM36003.1 | 9.99e-128 | 1 | 622 | 1 | 665 |
| VEJ07254.1 | 5.56e-127 | 1 | 622 | 1 | 665 |
| QOP40873.1 | 2.37e-53 | 14 | 622 | 19 | 698 |
| QFR48868.1 | 5.89e-53 | 14 | 622 | 18 | 720 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3RCE_A | 8.88e-34 | 2 | 622 | 12 | 710 | Bacterialoligosaccharyltransferase PglB [Campylobacter lari] |
| 5OGL_A | 2.73e-33 | 2 | 622 | 12 | 710 | Structureof bacterial oligosaccharyltransferase PglB in complex with an acceptor peptide and an lipid-linked oligosaccharide analog [Campylobacter lari RM2100] |
| 6GXC_A | 2.85e-33 | 2 | 622 | 12 | 710 | Bacterialoligosaccharyltransferase PglB in complex with an inhibitory peptide and a reactive lipid-linked oligosaccharide analog [Campylobacter lari RM2100] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0P9C8 | 7.55e-42 | 14 | 491 | 22 | 510 | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter jejuni subsp. jejuni serotype O:2 (strain ATCC 700819 / NCTC 11168) OX=192222 GN=pglB PE=1 SV=1 |
| Q5HTX9 | 1.87e-41 | 14 | 491 | 22 | 510 | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter jejuni (strain RM1221) OX=195099 GN=pglB PE=1 SV=1 |
| B9KDD4 | 1.93e-33 | 2 | 622 | 12 | 710 | Undecaprenyl-diphosphooligosaccharide--protein glycotransferase OS=Campylobacter lari (strain RM2100 / D67 / ATCC BAA-1060) OX=306263 GN=pglB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.982216 | 0.016608 | 0.000435 | 0.000075 | 0.000048 | 0.000640 |
TMHMM Annotations download full data without filtering help
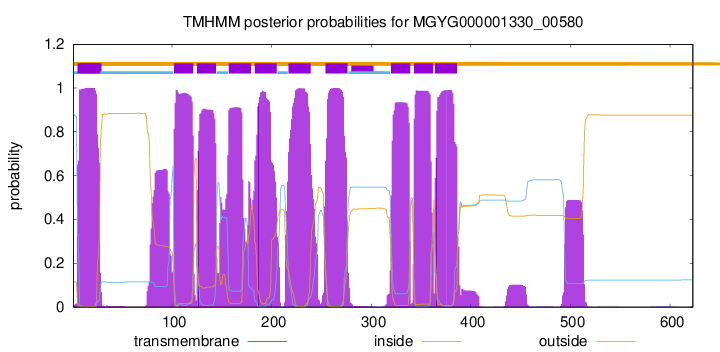
| start | end |
|---|---|
| 5 | 27 |
| 102 | 121 |
| 125 | 144 |
| 157 | 179 |
| 183 | 205 |
| 217 | 239 |
| 254 | 276 |
| 320 | 339 |
| 343 | 362 |
| 364 | 386 |
