You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001332_00606
Basic Information
help
| Species |
Massilioclostridium methylpentosum
|
| Lineage |
Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Massilioclostridium; Massilioclostridium methylpentosum
|
| CAZyme ID |
MGYG000001332_00606
|
| CAZy Family |
GH106 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1224 |
|
135144.81 |
4.1552 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000001332 |
3406391 |
Isolate |
not provided |
not provided |
|
| Gene Location |
Start: 681211;
End: 684885
Strand: -
|
No EC number prediction in MGYG000001332_00606.
| Family |
Start |
End |
Evalue |
family coverage |
| GH106 |
45 |
846 |
2.9e-73 |
0.8216019417475728 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam17132
|
Glyco_hydro_106 |
2.94e-15 |
206 |
708 |
338 |
805 |
alpha-L-rhamnosidase. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5MQM_A
|
1.80e-06 |
932 |
1038 |
968 |
1074 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A
|
1.80e-06 |
932 |
1038 |
968 |
1074 |
Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
This protein is predicted as SP
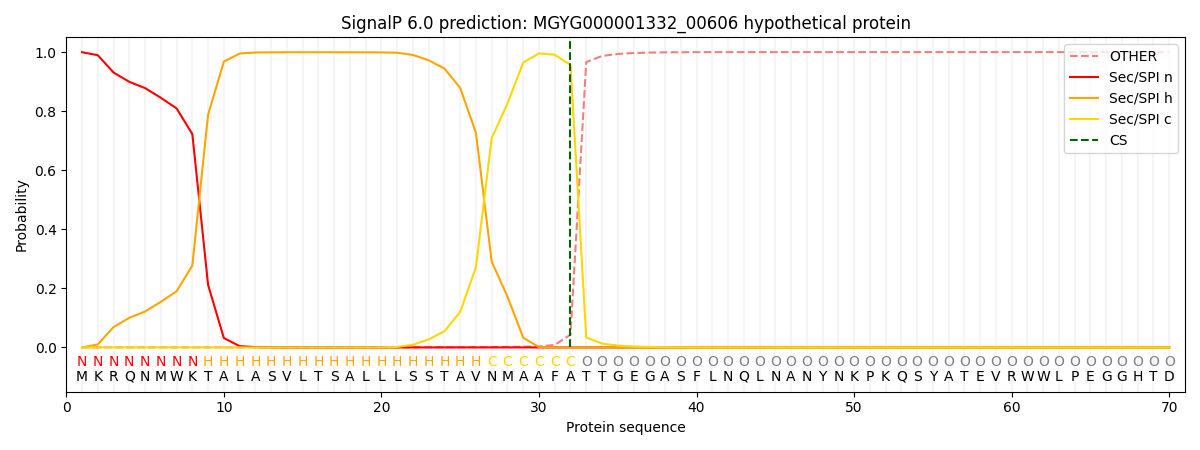
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000853
|
0.997712
|
0.000268
|
0.000647
|
0.000260
|
0.000217
|
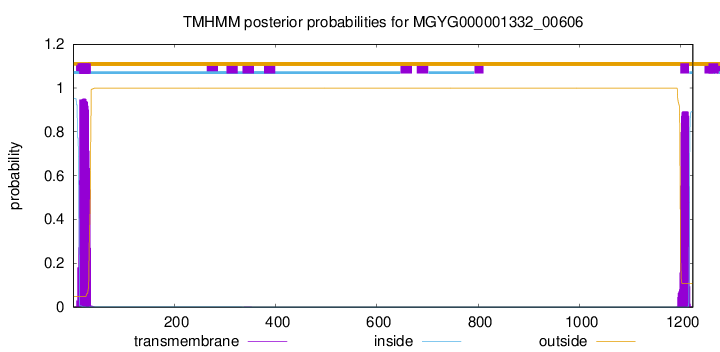
| start |
end |
| 13 |
35 |
| 1199 |
1216 |


